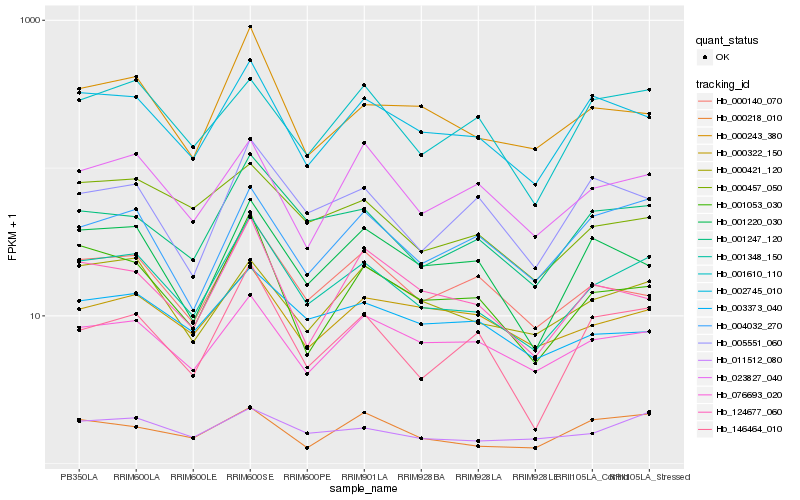
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001348_150 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At4g08330, chloroplastic [Jatropha curcas] |
| 2 |
Hb_001247_120 |
0.0890651918 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 3 [Jatropha curcas] |
| 3 |
Hb_002745_010 |
0.0964343248 |
- |
- |
PREDICTED: SUMO-conjugating enzyme SCE1 [Cucumis sativus] |
| 4 |
Hb_076693_020 |
0.1095453385 |
- |
- |
hypothetical protein CISIN_1g0121812mg, partial [Citrus sinensis] |
| 5 |
Hb_001053_030 |
0.1113269183 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_000140_070 |
0.1118008896 |
- |
- |
PREDICTED: uncharacterized protein LOC105642935 isoform X1 [Jatropha curcas] |
| 7 |
Hb_011512_080 |
0.1121437342 |
- |
- |
hypothetical protein JCGZ_06424 [Jatropha curcas] |
| 8 |
Hb_146464_010 |
0.1134632019 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_000243_380 |
0.1144653655 |
- |
- |
PREDICTED: histone H1 [Jatropha curcas] |
| 10 |
Hb_000421_120 |
0.1172268391 |
- |
- |
PREDICTED: NEDD8-specific protease 1 [Jatropha curcas] |
| 11 |
Hb_000457_050 |
0.1181184494 |
- |
- |
PREDICTED: tether containing UBX domain for GLUT4 [Jatropha curcas] |
| 12 |
Hb_001220_030 |
0.1189006403 |
- |
- |
PREDICTED: BRCA1-A complex subunit Abraxas [Jatropha curcas] |
| 13 |
Hb_001610_110 |
0.119582078 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 32 homolog 2 [Jatropha curcas] |
| 14 |
Hb_004032_270 |
0.1196176053 |
transcription factor |
TF Family: PLATZ |
protein with unknown function [Ricinus communis] |
| 15 |
Hb_124677_060 |
0.121662842 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 16 |
Hb_000218_010 |
0.1232404682 |
- |
- |
- |
| 17 |
Hb_023827_040 |
0.1239668011 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 11-like [Jatropha curcas] |
| 18 |
Hb_005551_060 |
0.1248424881 |
- |
- |
PREDICTED: uncharacterized protein LOC105635819 [Jatropha curcas] |
| 19 |
Hb_000322_150 |
0.1256800736 |
transcription factor |
TF Family: TRAF |
PREDICTED: regulatory protein NPR1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_003373_040 |
0.127957368 |
- |
- |
hypothetical protein ARALYDRAFT_356474 [Arabidopsis lyrata subsp. lyrata] |