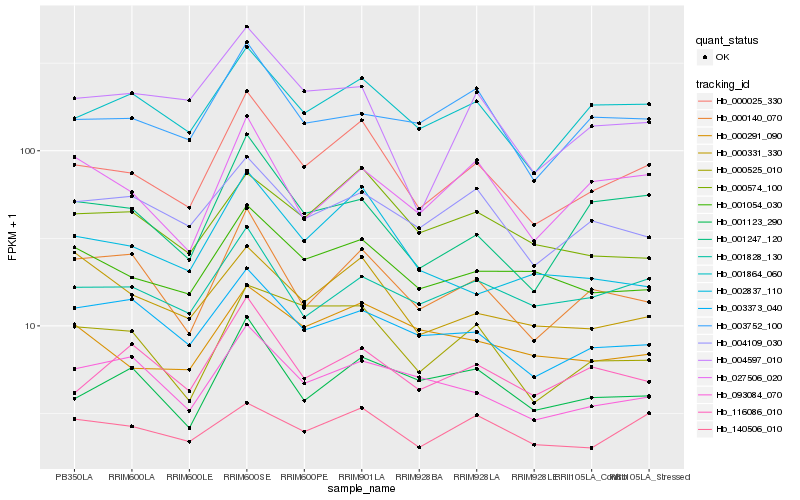
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000025_330 |
0.0 |
- |
- |
hypothetical protein JCGZ_22759 [Jatropha curcas] |
| 2 |
Hb_001864_060 |
0.0860735276 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1-like [Jatropha curcas] |
| 3 |
Hb_000140_070 |
0.1007463887 |
- |
- |
PREDICTED: uncharacterized protein LOC105642935 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001247_120 |
0.1062638803 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 3 [Jatropha curcas] |
| 5 |
Hb_001054_030 |
0.1067011994 |
transcription factor |
TF Family: NF-X1 |
PREDICTED: NF-X1-type zinc finger protein NFXL1 [Jatropha curcas] |
| 6 |
Hb_000525_010 |
0.1082967071 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 7 |
Hb_001123_290 |
0.1104490185 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 3 [Jatropha curcas] |
| 8 |
Hb_001828_130 |
0.1115711791 |
- |
- |
PREDICTED: transcription factor GTE8-like [Jatropha curcas] |
| 9 |
Hb_116086_010 |
0.1119572914 |
- |
- |
hypothetical protein JCGZ_14829 [Jatropha curcas] |
| 10 |
Hb_000574_100 |
0.1135133497 |
- |
- |
DEAD-box protein abstrakt [Populus trichocarpa] |
| 11 |
Hb_004109_030 |
0.114705056 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_027506_020 |
0.1155724198 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g30090 [Jatropha curcas] |
| 13 |
Hb_003752_100 |
0.1162359072 |
- |
- |
PREDICTED: FRIGIDA-like protein 3 [Jatropha curcas] |
| 14 |
Hb_002837_110 |
0.1162471295 |
- |
- |
PREDICTED: probable 3-hydroxyisobutyrate dehydrogenase-like 2, mitochondrial [Jatropha curcas] |
| 15 |
Hb_003373_040 |
0.1171799367 |
- |
- |
hypothetical protein ARALYDRAFT_356474 [Arabidopsis lyrata subsp. lyrata] |
| 16 |
Hb_093084_070 |
0.1185238763 |
desease resistance |
Gene Name: Baculo_PEP_C |
Disease resistance protein RPP13, putative [Ricinus communis] |
| 17 |
Hb_000291_090 |
0.1196118556 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 7 isoform X2 [Jatropha curcas] |
| 18 |
Hb_140506_010 |
0.1203226554 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g33170-like [Jatropha curcas] |
| 19 |
Hb_004597_010 |
0.1219379439 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor MYC2 [Jatropha curcas] |
| 20 |
Hb_000331_330 |
0.1227825222 |
- |
- |
PREDICTED: transcription factor GTE10 isoform X1 [Jatropha curcas] |