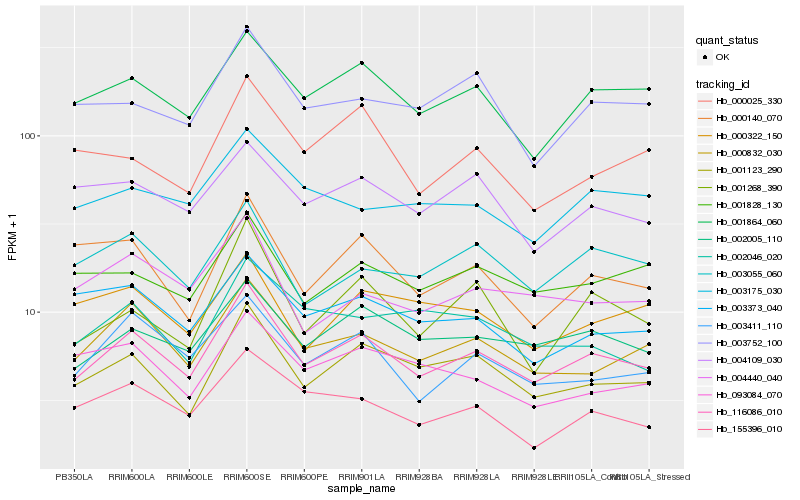
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_116086_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_14829 [Jatropha curcas] |
| 2 |
Hb_001123_290 |
0.0791996301 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 3 [Jatropha curcas] |
| 3 |
Hb_000832_030 |
0.0798868749 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105634915 [Jatropha curcas] |
| 4 |
Hb_001864_060 |
0.0811692418 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1-like [Jatropha curcas] |
| 5 |
Hb_002005_110 |
0.0816278885 |
- |
- |
PREDICTED: CTD small phosphatase-like protein 2 isoform X5 [Jatropha curcas] |
| 6 |
Hb_003411_110 |
0.0919797842 |
- |
- |
PREDICTED: uncharacterized protein LOC105642013 [Jatropha curcas] |
| 7 |
Hb_155396_010 |
0.0954325452 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 8 |
Hb_001828_130 |
0.1003776106 |
- |
- |
PREDICTED: transcription factor GTE8-like [Jatropha curcas] |
| 9 |
Hb_003055_060 |
0.1009171337 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105649487 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001268_390 |
0.1014772071 |
- |
- |
PREDICTED: uncharacterized protein LOC105108367 isoform X1 [Populus euphratica] |
| 11 |
Hb_003752_100 |
0.1025659511 |
- |
- |
PREDICTED: FRIGIDA-like protein 3 [Jatropha curcas] |
| 12 |
Hb_004440_040 |
0.1042038429 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 13 |
Hb_000140_070 |
0.1054643349 |
- |
- |
PREDICTED: uncharacterized protein LOC105642935 isoform X1 [Jatropha curcas] |
| 14 |
Hb_004109_030 |
0.1057952287 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_003175_030 |
0.1058496879 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410-like [Populus euphratica] |
| 16 |
Hb_003373_040 |
0.1103122808 |
- |
- |
hypothetical protein ARALYDRAFT_356474 [Arabidopsis lyrata subsp. lyrata] |
| 17 |
Hb_000025_330 |
0.1119572914 |
- |
- |
hypothetical protein JCGZ_22759 [Jatropha curcas] |
| 18 |
Hb_000322_150 |
0.1133774337 |
transcription factor |
TF Family: TRAF |
PREDICTED: regulatory protein NPR1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002046_020 |
0.1148031992 |
- |
- |
PREDICTED: putative clathrin assembly protein At2g25430 [Jatropha curcas] |
| 20 |
Hb_093084_070 |
0.1149751941 |
desease resistance |
Gene Name: Baculo_PEP_C |
Disease resistance protein RPP13, putative [Ricinus communis] |