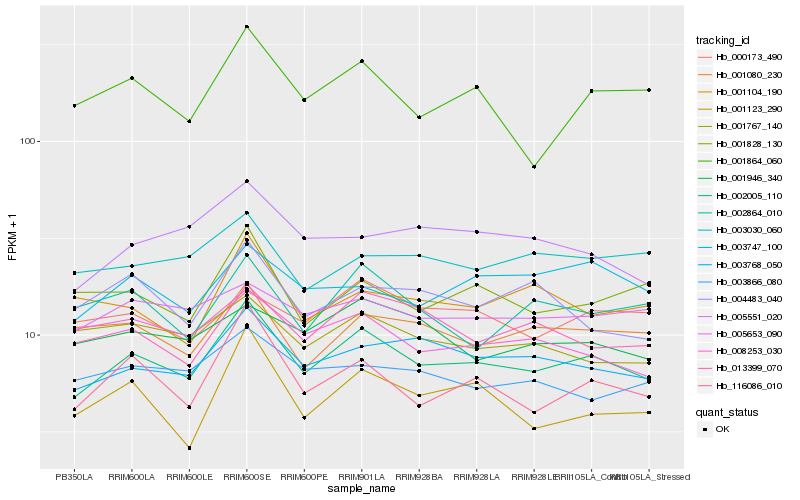
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002005_110 |
0.0 |
- |
- |
PREDICTED: CTD small phosphatase-like protein 2 isoform X5 [Jatropha curcas] |
| 2 |
Hb_003768_050 |
0.0783043621 |
- |
- |
PREDICTED: uncharacterized protein LOC105644152 isoform X1 [Jatropha curcas] |
| 3 |
Hb_116086_010 |
0.0816278885 |
- |
- |
hypothetical protein JCGZ_14829 [Jatropha curcas] |
| 4 |
Hb_013399_070 |
0.0923121999 |
- |
- |
PREDICTED: uncharacterized protein LOC105631409 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001864_060 |
0.0948299152 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1-like [Jatropha curcas] |
| 6 |
Hb_001080_230 |
0.0979391587 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105649979 [Jatropha curcas] |
| 7 |
Hb_001828_130 |
0.0981661635 |
- |
- |
PREDICTED: transcription factor GTE8-like [Jatropha curcas] |
| 8 |
Hb_001123_290 |
0.0990087955 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 3 [Jatropha curcas] |
| 9 |
Hb_005551_020 |
0.1013721485 |
- |
- |
PREDICTED: probable receptor protein kinase TMK1 [Jatropha curcas] |
| 10 |
Hb_003747_100 |
0.101981778 |
- |
- |
PREDICTED: probable tyrosine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 11 |
Hb_005653_090 |
0.102256293 |
- |
- |
PREDICTED: protein HIRA isoform X1 [Jatropha curcas] |
| 12 |
Hb_002864_010 |
0.1035031756 |
- |
- |
PREDICTED: regulator of nonsense transcripts 1 homolog [Jatropha curcas] |
| 13 |
Hb_001767_140 |
0.103665545 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase MKP1 [Jatropha curcas] |
| 14 |
Hb_003866_080 |
0.1043260334 |
- |
- |
PREDICTED: uncharacterized protein LOC105639150 [Jatropha curcas] |
| 15 |
Hb_008253_030 |
0.1050087713 |
- |
- |
PREDICTED: probable carboxylesterase 11 [Jatropha curcas] |
| 16 |
Hb_001946_340 |
0.1050462735 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 17-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_004483_040 |
0.1051738776 |
- |
- |
hypothetical protein JCGZ_22047 [Jatropha curcas] |
| 18 |
Hb_003030_060 |
0.1054561719 |
- |
- |
PREDICTED: regulation of nuclear pre-mRNA domain-containing protein 2-like [Jatropha curcas] |
| 19 |
Hb_000173_490 |
0.1062535879 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 4 isoform X2 [Jatropha curcas] |
| 20 |
Hb_001104_190 |
0.106552336 |
- |
- |
PREDICTED: uncharacterized protein LOC105629451 [Jatropha curcas] |