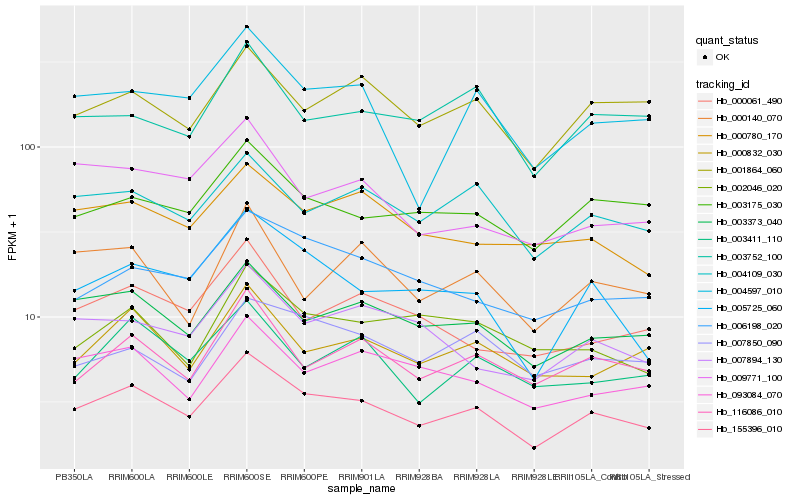
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_155396_010 |
0.0 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 2 |
Hb_003373_040 |
0.0824350382 |
- |
- |
hypothetical protein ARALYDRAFT_356474 [Arabidopsis lyrata subsp. lyrata] |
| 3 |
Hb_004109_030 |
0.0941952047 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_116086_010 |
0.0954325452 |
- |
- |
hypothetical protein JCGZ_14829 [Jatropha curcas] |
| 5 |
Hb_005725_060 |
0.0975702161 |
- |
- |
PREDICTED: U-box domain-containing protein 30-like [Jatropha curcas] |
| 6 |
Hb_001864_060 |
0.0989233549 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1-like [Jatropha curcas] |
| 7 |
Hb_003175_030 |
0.099545072 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410-like [Populus euphratica] |
| 8 |
Hb_000832_030 |
0.1014480404 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105634915 [Jatropha curcas] |
| 9 |
Hb_006198_020 |
0.1017639903 |
- |
- |
PREDICTED: uncharacterized protein LOC105644416 [Jatropha curcas] |
| 10 |
Hb_093084_070 |
0.1022000733 |
desease resistance |
Gene Name: Baculo_PEP_C |
Disease resistance protein RPP13, putative [Ricinus communis] |
| 11 |
Hb_000780_170 |
0.1049378378 |
- |
- |
PREDICTED: OBERON-like protein [Jatropha curcas] |
| 12 |
Hb_004597_010 |
0.1059765445 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor MYC2 [Jatropha curcas] |
| 13 |
Hb_003411_110 |
0.1081258822 |
- |
- |
PREDICTED: uncharacterized protein LOC105642013 [Jatropha curcas] |
| 14 |
Hb_000061_490 |
0.1083991069 |
- |
- |
hypothetical protein CISIN_1g0362002mg, partial [Citrus sinensis] |
| 15 |
Hb_007894_130 |
0.1099058229 |
- |
- |
hypothetical protein RCOM_0584960 [Ricinus communis] |
| 16 |
Hb_003752_100 |
0.1106857617 |
- |
- |
PREDICTED: FRIGIDA-like protein 3 [Jatropha curcas] |
| 17 |
Hb_009771_100 |
0.1134289171 |
- |
- |
PREDICTED: uncharacterized protein LOC105141784 [Populus euphratica] |
| 18 |
Hb_007850_090 |
0.1138536945 |
- |
- |
PREDICTED: probable galacturonosyltransferase 11 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002046_020 |
0.1143530205 |
- |
- |
PREDICTED: putative clathrin assembly protein At2g25430 [Jatropha curcas] |
| 20 |
Hb_000140_070 |
0.1153329027 |
- |
- |
PREDICTED: uncharacterized protein LOC105642935 isoform X1 [Jatropha curcas] |