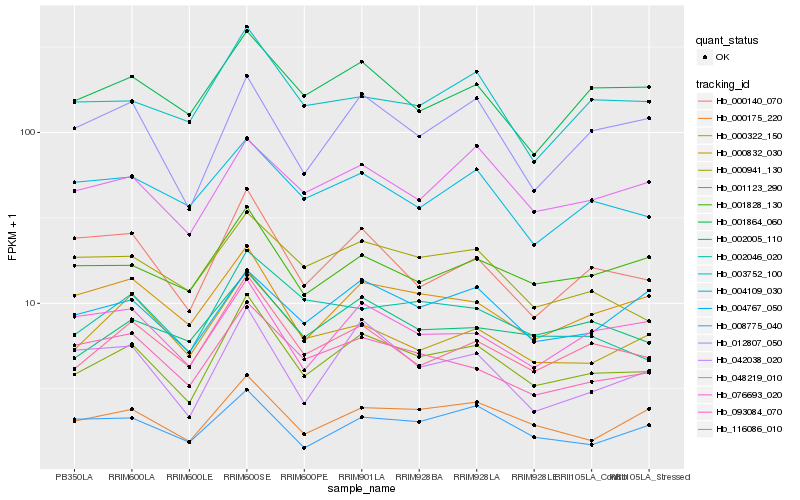
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001123_290 |
0.0 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 3 [Jatropha curcas] |
| 2 |
Hb_116086_010 |
0.0791996301 |
- |
- |
hypothetical protein JCGZ_14829 [Jatropha curcas] |
| 3 |
Hb_000175_220 |
0.0910485705 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g21065-like [Jatropha curcas] |
| 4 |
Hb_000322_150 |
0.0959886076 |
transcription factor |
TF Family: TRAF |
PREDICTED: regulatory protein NPR1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000832_030 |
0.0984539597 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105634915 [Jatropha curcas] |
| 6 |
Hb_001864_060 |
0.0990047287 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1-like [Jatropha curcas] |
| 7 |
Hb_002005_110 |
0.0990087955 |
- |
- |
PREDICTED: CTD small phosphatase-like protein 2 isoform X5 [Jatropha curcas] |
| 8 |
Hb_093084_070 |
0.0997054652 |
desease resistance |
Gene Name: Baculo_PEP_C |
Disease resistance protein RPP13, putative [Ricinus communis] |
| 9 |
Hb_000140_070 |
0.0999395298 |
- |
- |
PREDICTED: uncharacterized protein LOC105642935 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001828_130 |
0.1009516348 |
- |
- |
PREDICTED: transcription factor GTE8-like [Jatropha curcas] |
| 11 |
Hb_012807_050 |
0.1021459431 |
desease resistance |
Gene Name: CDC48_N |
PREDICTED: cell division cycle protein 48 homolog [Jatropha curcas] |
| 12 |
Hb_002046_020 |
0.1024586187 |
- |
- |
PREDICTED: putative clathrin assembly protein At2g25430 [Jatropha curcas] |
| 13 |
Hb_003752_100 |
0.1030343237 |
- |
- |
PREDICTED: FRIGIDA-like protein 3 [Jatropha curcas] |
| 14 |
Hb_048219_010 |
0.1042551779 |
- |
- |
coronatine-insensitive 1 [Hevea brasiliensis] |
| 15 |
Hb_000941_130 |
0.1068542758 |
- |
- |
gamma-tubulin complex component, putative [Ricinus communis] |
| 16 |
Hb_008775_040 |
0.1070841697 |
- |
- |
Pentatricopeptide repeat (PPR) superfamily protein, putative [Theobroma cacao] |
| 17 |
Hb_004767_050 |
0.1074130531 |
- |
- |
PREDICTED: glycosyltransferase family protein 64 C3-like [Jatropha curcas] |
| 18 |
Hb_042038_020 |
0.107640303 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g05670, mitochondrial [Jatropha curcas] |
| 19 |
Hb_076693_020 |
0.1077223387 |
- |
- |
hypothetical protein CISIN_1g0121812mg, partial [Citrus sinensis] |
| 20 |
Hb_004109_030 |
0.1088447115 |
- |
- |
conserved hypothetical protein [Ricinus communis] |