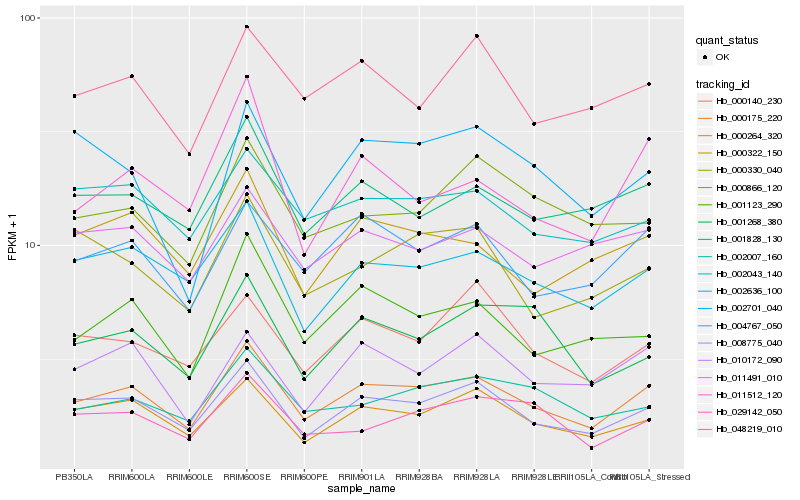
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000175_220 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g21065-like [Jatropha curcas] |
| 2 |
Hb_008775_040 |
0.0611745566 |
- |
- |
Pentatricopeptide repeat (PPR) superfamily protein, putative [Theobroma cacao] |
| 3 |
Hb_000264_320 |
0.0778541135 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g33760 [Jatropha curcas] |
| 4 |
Hb_004767_050 |
0.0844341086 |
- |
- |
PREDICTED: glycosyltransferase family protein 64 C3-like [Jatropha curcas] |
| 5 |
Hb_002636_100 |
0.0867319336 |
- |
- |
PREDICTED: DNA-directed RNA polymerase 1B, mitochondrial [Jatropha curcas] |
| 6 |
Hb_001123_290 |
0.0910485705 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 3 [Jatropha curcas] |
| 7 |
Hb_001828_130 |
0.0948403531 |
- |
- |
PREDICTED: transcription factor GTE8-like [Jatropha curcas] |
| 8 |
Hb_002007_160 |
0.0957291361 |
- |
- |
PREDICTED: uncharacterized protein LOC105647522 [Jatropha curcas] |
| 9 |
Hb_000322_150 |
0.0961486001 |
transcription factor |
TF Family: TRAF |
PREDICTED: regulatory protein NPR1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_048219_010 |
0.099830907 |
- |
- |
coronatine-insensitive 1 [Hevea brasiliensis] |
| 11 |
Hb_002043_140 |
0.0999557931 |
- |
- |
PREDICTED: uncharacterized protein At1g51745-like [Jatropha curcas] |
| 12 |
Hb_010172_090 |
0.1019861628 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_000330_040 |
0.1041061412 |
- |
- |
PREDICTED: protein RER1B-like [Jatropha curcas] |
| 14 |
Hb_001268_380 |
0.1051834875 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g14080-like [Jatropha curcas] |
| 15 |
Hb_011491_010 |
0.1052646241 |
- |
- |
Protein YME1, putative [Ricinus communis] |
| 16 |
Hb_000140_230 |
0.1052700414 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g61520, mitochondrial-like [Jatropha curcas] |
| 17 |
Hb_011512_120 |
0.1055872858 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 1 [Jatropha curcas] |
| 18 |
Hb_000866_120 |
0.1064339233 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 12 [Jatropha curcas] |
| 19 |
Hb_002701_040 |
0.1068639974 |
- |
- |
PREDICTED: auxin transport protein BIG [Jatropha curcas] |
| 20 |
Hb_029142_050 |
0.1073540072 |
desease resistance |
Gene Name: PPR |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |