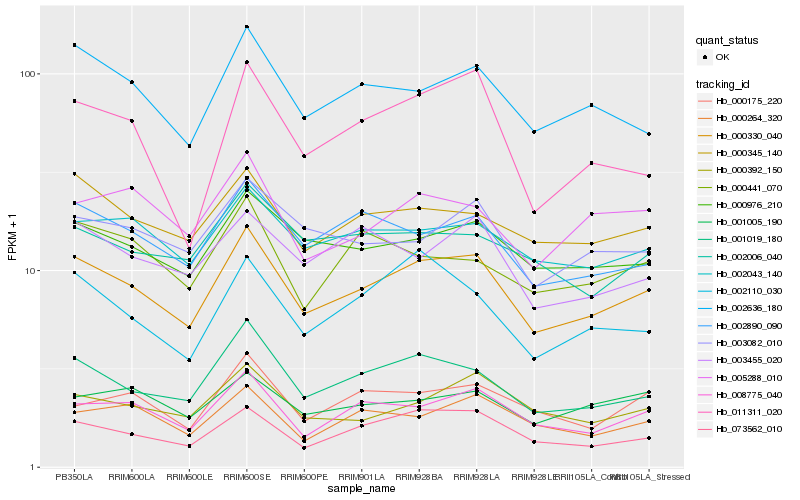
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000330_040 |
0.0 |
- |
- |
PREDICTED: protein RER1B-like [Jatropha curcas] |
| 2 |
Hb_001019_180 |
0.0717177549 |
- |
- |
PREDICTED: uncharacterized protein LOC105639324 [Jatropha curcas] |
| 3 |
Hb_073562_010 |
0.0797637039 |
- |
- |
hypothetical protein POPTR_0011s036602g, partial [Populus trichocarpa] |
| 4 |
Hb_000976_210 |
0.0808809249 |
- |
- |
PREDICTED: protein PAT1 homolog 1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_008775_040 |
0.0819500641 |
- |
- |
Pentatricopeptide repeat (PPR) superfamily protein, putative [Theobroma cacao] |
| 6 |
Hb_002636_180 |
0.0821316102 |
- |
- |
inositol or phosphatidylinositol kinase, putative [Ricinus communis] |
| 7 |
Hb_000345_140 |
0.0827446454 |
- |
- |
PREDICTED: autophagy-related protein 18a-like [Jatropha curcas] |
| 8 |
Hb_002890_090 |
0.0829547755 |
- |
- |
Auxin response factor 1 [Glycine soja] |
| 9 |
Hb_002043_140 |
0.0850720518 |
- |
- |
PREDICTED: uncharacterized protein At1g51745-like [Jatropha curcas] |
| 10 |
Hb_000392_150 |
0.0909537003 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g74580 [Jatropha curcas] |
| 11 |
Hb_003082_010 |
0.0934894411 |
- |
- |
PREDICTED: putative F-box protein At3g10240 [Jatropha curcas] |
| 12 |
Hb_002110_030 |
0.0939811916 |
- |
- |
PREDICTED: WD and tetratricopeptide repeats protein 1 isoform X3 [Jatropha curcas] |
| 13 |
Hb_005288_010 |
0.0971233854 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 14 |
Hb_001005_190 |
0.0973508613 |
- |
- |
PREDICTED: UPF0496 protein At3g19330-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_003455_020 |
0.0983735977 |
- |
- |
PREDICTED: transportin-1 [Jatropha curcas] |
| 16 |
Hb_002006_040 |
0.0999837607 |
- |
- |
PREDICTED: translation initiation factor eIF-2B subunit delta isoform X1 [Jatropha curcas] |
| 17 |
Hb_000264_320 |
0.100171053 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g33760 [Jatropha curcas] |
| 18 |
Hb_000441_070 |
0.1027677274 |
transcription factor |
TF Family: C3H |
PREDICTED: 30-kDa cleavage and polyadenylation specificity factor 30 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000175_220 |
0.1041061412 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g21065-like [Jatropha curcas] |
| 20 |
Hb_011311_020 |
0.1052447921 |
- |
- |
PREDICTED: myosin-11 isoform X2 [Jatropha curcas] |