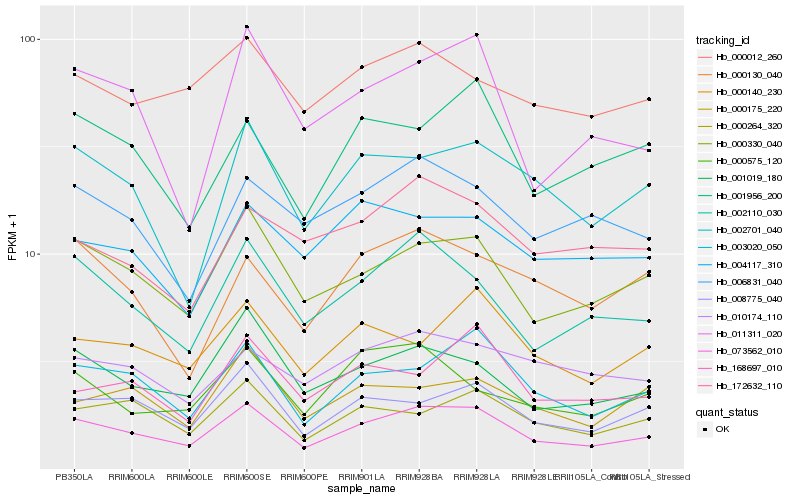
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_073562_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0011s036602g, partial [Populus trichocarpa] |
| 2 |
Hb_000330_040 |
0.0797637039 |
- |
- |
PREDICTED: protein RER1B-like [Jatropha curcas] |
| 3 |
Hb_003020_050 |
0.0847838511 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 4 |
Hb_002110_030 |
0.090532315 |
- |
- |
PREDICTED: WD and tetratricopeptide repeats protein 1 isoform X3 [Jatropha curcas] |
| 5 |
Hb_008775_040 |
0.0963105531 |
- |
- |
Pentatricopeptide repeat (PPR) superfamily protein, putative [Theobroma cacao] |
| 6 |
Hb_001019_180 |
0.0997852969 |
- |
- |
PREDICTED: uncharacterized protein LOC105639324 [Jatropha curcas] |
| 7 |
Hb_002701_040 |
0.1046420491 |
- |
- |
PREDICTED: auxin transport protein BIG [Jatropha curcas] |
| 8 |
Hb_000264_320 |
0.1082893043 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g33760 [Jatropha curcas] |
| 9 |
Hb_011311_020 |
0.109265183 |
- |
- |
PREDICTED: myosin-11 isoform X2 [Jatropha curcas] |
| 10 |
Hb_001956_200 |
0.1108146478 |
- |
- |
pumilio/Puf RNA-binding domain-containing family protein [Populus trichocarpa] |
| 11 |
Hb_000140_230 |
0.1116510093 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g61520, mitochondrial-like [Jatropha curcas] |
| 12 |
Hb_006831_040 |
0.1134660568 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_010174_110 |
0.1159099044 |
- |
- |
hypothetical protein JCGZ_17880 [Jatropha curcas] |
| 14 |
Hb_000130_040 |
0.119649451 |
- |
- |
PREDICTED: dol-P-Man:Man(6)GlcNAc(2)-PP-Dol alpha-1,2-mannosyltransferase [Jatropha curcas] |
| 15 |
Hb_000012_260 |
0.1199986006 |
- |
- |
Far upstream element-binding 2 [Gossypium arboreum] |
| 16 |
Hb_168697_010 |
0.1207341212 |
- |
- |
hypothetical protein JCGZ_16772 [Jatropha curcas] |
| 17 |
Hb_004117_310 |
0.1213101828 |
- |
- |
PREDICTED: uncharacterized protein LOC105648043 [Jatropha curcas] |
| 18 |
Hb_000175_220 |
0.121856371 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g21065-like [Jatropha curcas] |
| 19 |
Hb_172632_110 |
0.1223820227 |
- |
- |
PREDICTED: pumilio homolog 5 [Jatropha curcas] |
| 20 |
Hb_000575_120 |
0.1223906233 |
- |
- |
hypothetical protein POPTR_0011s02060g [Populus trichocarpa] |