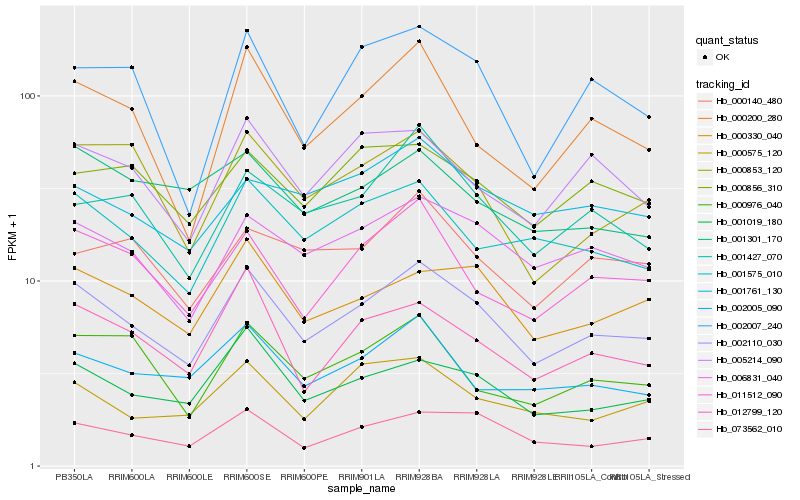
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002110_030 |
0.0 |
- |
- |
PREDICTED: WD and tetratricopeptide repeats protein 1 isoform X3 [Jatropha curcas] |
| 2 |
Hb_006831_040 |
0.0858182593 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_001019_180 |
0.0887942027 |
- |
- |
PREDICTED: uncharacterized protein LOC105639324 [Jatropha curcas] |
| 4 |
Hb_073562_010 |
0.090532315 |
- |
- |
hypothetical protein POPTR_0011s036602g, partial [Populus trichocarpa] |
| 5 |
Hb_000330_040 |
0.0939811916 |
- |
- |
PREDICTED: protein RER1B-like [Jatropha curcas] |
| 6 |
Hb_011512_090 |
0.0955808593 |
- |
- |
Hydroxyethylthiazole kinase, putative [Ricinus communis] |
| 7 |
Hb_001575_010 |
0.096430502 |
- |
- |
PREDICTED: pumilio homolog 6, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_012799_120 |
0.1012777135 |
- |
- |
PREDICTED: DNA mismatch repair protein MSH3 isoform X1 [Jatropha curcas] |
| 9 |
Hb_005214_090 |
0.1042575849 |
- |
- |
PREDICTED: uncharacterized protein LOC105636015 [Jatropha curcas] |
| 10 |
Hb_002005_090 |
0.1048861787 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000200_280 |
0.1051934352 |
- |
- |
PREDICTED: uncharacterized protein LOC105636924 [Jatropha curcas] |
| 12 |
Hb_001427_070 |
0.1077572267 |
- |
- |
PREDICTED: xaa-Pro dipeptidase [Jatropha curcas] |
| 13 |
Hb_000853_120 |
0.1079179169 |
- |
- |
PREDICTED: ethylene response sensor 1 [Jatropha curcas] |
| 14 |
Hb_000575_120 |
0.1080294601 |
- |
- |
hypothetical protein POPTR_0011s02060g [Populus trichocarpa] |
| 15 |
Hb_001761_130 |
0.112808513 |
- |
- |
PREDICTED: uncharacterized protein LOC105646118 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000856_310 |
0.1128186325 |
- |
- |
PREDICTED: hypersensitive-induced response protein 1 [Jatropha curcas] |
| 17 |
Hb_002007_240 |
0.1129466315 |
- |
- |
PREDICTED: F-box protein At1g55000 [Jatropha curcas] |
| 18 |
Hb_000976_040 |
0.1153487527 |
- |
- |
molybdopterin cofactor sulfurase, putative [Ricinus communis] |
| 19 |
Hb_000140_480 |
0.1176328698 |
- |
- |
PREDICTED: uncharacterized protein LOC105637350 [Jatropha curcas] |
| 20 |
Hb_001301_170 |
0.1179822018 |
- |
- |
PREDICTED: purple acid phosphatase 18 [Jatropha curcas] |