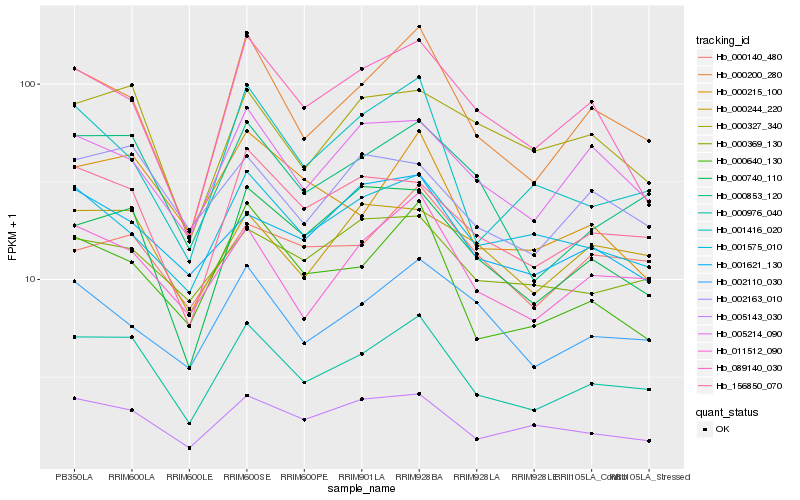
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000976_040 |
0.0 |
- |
- |
molybdopterin cofactor sulfurase, putative [Ricinus communis] |
| 2 |
Hb_000853_120 |
0.0801972594 |
- |
- |
PREDICTED: ethylene response sensor 1 [Jatropha curcas] |
| 3 |
Hb_000200_280 |
0.0829968564 |
- |
- |
PREDICTED: uncharacterized protein LOC105636924 [Jatropha curcas] |
| 4 |
Hb_011512_090 |
0.0863360033 |
- |
- |
Hydroxyethylthiazole kinase, putative [Ricinus communis] |
| 5 |
Hb_156850_070 |
0.0950687447 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000640_130 |
0.0998064644 |
transcription factor |
TF Family: SET |
unnamed protein product [Vitis vinifera] |
| 7 |
Hb_005143_030 |
0.1017351326 |
- |
- |
PREDICTED: uncharacterized protein LOC105638843 [Jatropha curcas] |
| 8 |
Hb_000740_110 |
0.1080686095 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_01850 [Jatropha curcas] |
| 9 |
Hb_005214_090 |
0.1090120671 |
- |
- |
PREDICTED: uncharacterized protein LOC105636015 [Jatropha curcas] |
| 10 |
Hb_000244_220 |
0.1103513746 |
- |
- |
PREDICTED: bifunctional aspartokinase/homoserine dehydrogenase 1, chloroplastic-like [Jatropha curcas] |
| 11 |
Hb_002163_010 |
0.1112787599 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410 [Jatropha curcas] |
| 12 |
Hb_001575_010 |
0.1129715421 |
- |
- |
PREDICTED: pumilio homolog 6, chloroplastic isoform X1 [Jatropha curcas] |
| 13 |
Hb_000327_340 |
0.1152822848 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360 [Jatropha curcas] |
| 14 |
Hb_002110_030 |
0.1153487527 |
- |
- |
PREDICTED: WD and tetratricopeptide repeats protein 1 isoform X3 [Jatropha curcas] |
| 15 |
Hb_001416_020 |
0.1170000854 |
- |
- |
H(\+)-transporting atpase plant/fungi plasma membrane type, putative [Ricinus communis] |
| 16 |
Hb_000215_100 |
0.1177156448 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000369_130 |
0.1191183094 |
- |
- |
PREDICTED: H/ACA ribonucleoprotein complex subunit 4 [Jatropha curcas] |
| 18 |
Hb_001621_130 |
0.1191545726 |
- |
- |
PREDICTED: pumilio homolog 1-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_000140_480 |
0.1197328795 |
- |
- |
PREDICTED: uncharacterized protein LOC105637350 [Jatropha curcas] |
| 20 |
Hb_089140_030 |
0.1205941768 |
- |
- |
PREDICTED: thiol protease aleurain-like [Jatropha curcas] |