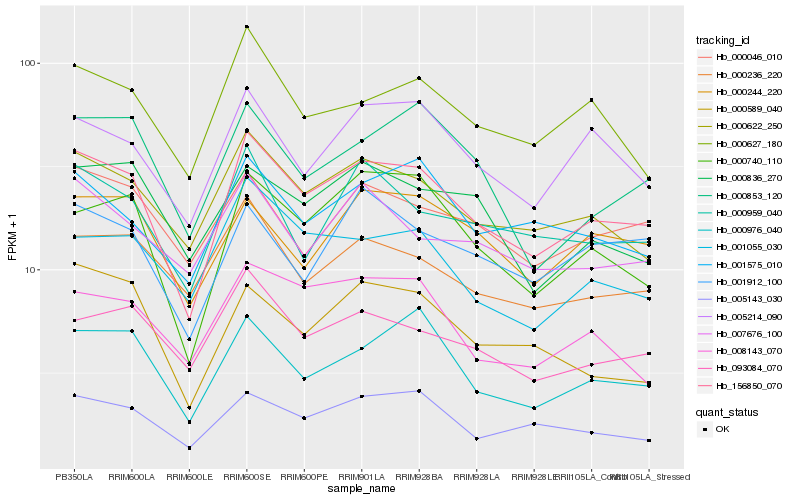
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_156850_070 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000622_250 |
0.0843479679 |
- |
- |
atpob1, putative [Ricinus communis] |
| 3 |
Hb_000740_110 |
0.0922429176 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_01850 [Jatropha curcas] |
| 4 |
Hb_000976_040 |
0.0950687447 |
- |
- |
molybdopterin cofactor sulfurase, putative [Ricinus communis] |
| 5 |
Hb_005143_030 |
0.0959081266 |
- |
- |
PREDICTED: uncharacterized protein LOC105638843 [Jatropha curcas] |
| 6 |
Hb_005214_090 |
0.0979323087 |
- |
- |
PREDICTED: uncharacterized protein LOC105636015 [Jatropha curcas] |
| 7 |
Hb_000627_180 |
0.1028242086 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_093084_070 |
0.1040343412 |
desease resistance |
Gene Name: Baculo_PEP_C |
Disease resistance protein RPP13, putative [Ricinus communis] |
| 9 |
Hb_000959_040 |
0.1041005806 |
- |
- |
PREDICTED: uncharacterized protein LOC105636570 [Jatropha curcas] |
| 10 |
Hb_001575_010 |
0.104932218 |
- |
- |
PREDICTED: pumilio homolog 6, chloroplastic isoform X1 [Jatropha curcas] |
| 11 |
Hb_000236_220 |
0.105973183 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-5 [Jatropha curcas] |
| 12 |
Hb_000244_220 |
0.1066012861 |
- |
- |
PREDICTED: bifunctional aspartokinase/homoserine dehydrogenase 1, chloroplastic-like [Jatropha curcas] |
| 13 |
Hb_001055_030 |
0.1068644425 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 14 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000046_010 |
0.1087053245 |
- |
- |
PREDICTED: histidine-containing phosphotransfer protein 1 [Jatropha curcas] |
| 15 |
Hb_001912_100 |
0.1089191916 |
- |
- |
PREDICTED: uncharacterized protein LOC105636609 [Jatropha curcas] |
| 16 |
Hb_000853_120 |
0.1090176004 |
- |
- |
PREDICTED: ethylene response sensor 1 [Jatropha curcas] |
| 17 |
Hb_000589_040 |
0.1106356526 |
desease resistance |
Gene Name: zf-BED |
PREDICTED: probable disease resistance protein At1g12280 [Jatropha curcas] |
| 18 |
Hb_008143_070 |
0.1112312133 |
- |
- |
PREDICTED: factor of DNA methylation 4 [Jatropha curcas] |
| 19 |
Hb_007676_100 |
0.1132922141 |
- |
- |
PREDICTED: uncharacterized protein LOC105646857 [Jatropha curcas] |
| 20 |
Hb_000836_270 |
0.1134595343 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog MMK2 [Jatropha curcas] |