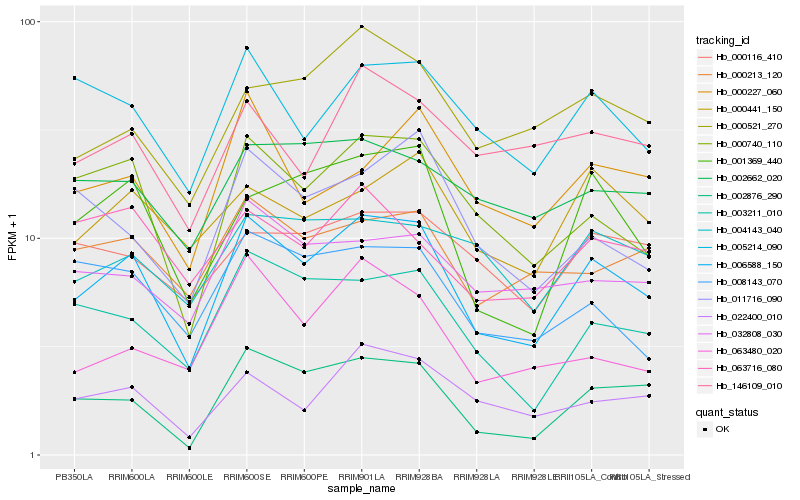
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006588_150 |
0.0 |
- |
- |
PREDICTED: steroid 5-alpha-reductase DET2 [Jatropha curcas] |
| 2 |
Hb_000740_110 |
0.1101419802 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_01850 [Jatropha curcas] |
| 3 |
Hb_002876_290 |
0.1178188611 |
- |
- |
PREDICTED: uncharacterized protein LOC104446353 [Eucalyptus grandis] |
| 4 |
Hb_001369_440 |
0.1194137969 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 7 [Jatropha curcas] |
| 5 |
Hb_003211_010 |
0.1209935637 |
- |
- |
PREDICTED: uncharacterized protein LOC105635705 [Jatropha curcas] |
| 6 |
Hb_022400_010 |
0.124250992 |
- |
- |
Nitrilase/cyanide hydratase and apolipoprotein N-acyltransferase family protein [Theobroma cacao] |
| 7 |
Hb_005214_090 |
0.1296619976 |
- |
- |
PREDICTED: uncharacterized protein LOC105636015 [Jatropha curcas] |
| 8 |
Hb_000213_120 |
0.1335655232 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 9 |
Hb_000521_270 |
0.1345398262 |
- |
- |
PREDICTED: protein EMSY-LIKE 3 [Jatropha curcas] |
| 10 |
Hb_000441_150 |
0.1366660936 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, root-type isozyme, chloroplastic isoform X2 [Jatropha curcas] |
| 11 |
Hb_008143_070 |
0.1369778451 |
- |
- |
PREDICTED: factor of DNA methylation 4 [Jatropha curcas] |
| 12 |
Hb_032808_030 |
0.1375528154 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 13 |
Hb_000227_060 |
0.1406500215 |
transcription factor |
TF Family: NF-YC |
hypothetical protein POPTR_0010s03730g [Populus trichocarpa] |
| 14 |
Hb_146109_010 |
0.1420751953 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002662_020 |
0.1427807193 |
- |
- |
PREDICTED: F-box protein At1g47056-like [Jatropha curcas] |
| 16 |
Hb_011716_090 |
0.1428252627 |
transcription factor |
TF Family: HRT |
putative transciption factor family protein [Populus trichocarpa] |
| 17 |
Hb_063480_020 |
0.1442778047 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_004143_040 |
0.1450730459 |
- |
- |
argininosuccinate lyase, putative [Ricinus communis] |
| 19 |
Hb_000116_410 |
0.1467535506 |
- |
- |
hypothetical protein JCGZ_22416 [Jatropha curcas] |
| 20 |
Hb_063716_080 |
0.1475365566 |
- |
- |
DNA-directed RNA polymerase III subunit F, putative [Ricinus communis] |