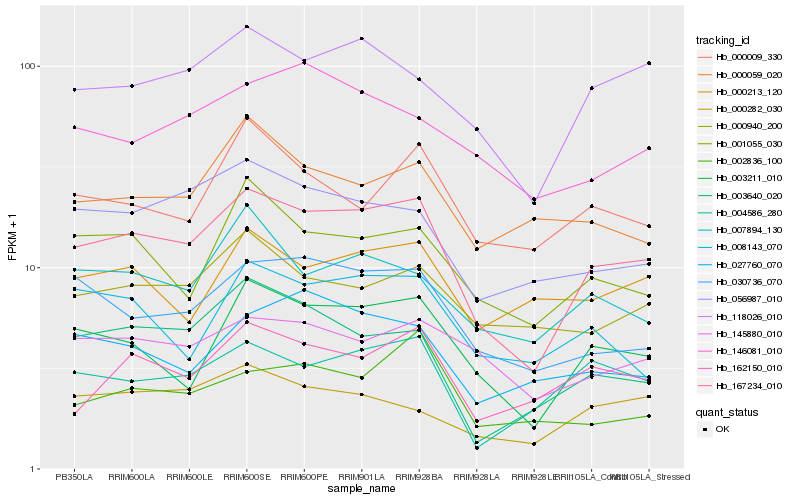
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_167234_010 |
0.0 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 13-like [Jatropha curcas] |
| 2 |
Hb_003211_010 |
0.1042349052 |
- |
- |
PREDICTED: uncharacterized protein LOC105635705 [Jatropha curcas] |
| 3 |
Hb_118026_010 |
0.1114629153 |
transcription factor |
TF Family: GNAT |
PREDICTED: N-alpha-acetyltransferase 11 [Jatropha curcas] |
| 4 |
Hb_030736_070 |
0.1156431558 |
- |
- |
PREDICTED: general transcription factor IIH subunit 3 isoform X2 [Jatropha curcas] |
| 5 |
Hb_056987_010 |
0.1222408457 |
- |
- |
DNA-damage-repair/toleration protein DRT111, chloroplastic [Glycine soja] |
| 6 |
Hb_004586_280 |
0.1237148853 |
- |
- |
PREDICTED: GEM-like protein 1 [Jatropha curcas] |
| 7 |
Hb_001055_030 |
0.1244760255 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 14 isoform X1 [Jatropha curcas] |
| 8 |
Hb_003640_020 |
0.1279847397 |
- |
- |
pelota, putative [Ricinus communis] |
| 9 |
Hb_007894_130 |
0.1280283734 |
- |
- |
hypothetical protein RCOM_0584960 [Ricinus communis] |
| 10 |
Hb_000282_030 |
0.1288428344 |
- |
- |
K(+) efflux antiporter 4 [Glycine soja] |
| 11 |
Hb_145880_010 |
0.128938226 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 12 |
Hb_162150_010 |
0.1297581211 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000940_200 |
0.1301366633 |
- |
- |
PREDICTED: uncharacterized protein LOC105643779 [Jatropha curcas] |
| 14 |
Hb_000009_330 |
0.1353188181 |
transcription factor |
TF Family: LIM |
zinc ion binding protein, putative [Ricinus communis] |
| 15 |
Hb_027760_070 |
0.1360749501 |
- |
- |
PREDICTED: uncharacterized endoplasmic reticulum membrane protein C16E8.02-like [Jatropha curcas] |
| 16 |
Hb_146081_010 |
0.1370372534 |
- |
- |
PREDICTED: protein SCAI [Jatropha curcas] |
| 17 |
Hb_008143_070 |
0.1387602495 |
- |
- |
PREDICTED: factor of DNA methylation 4 [Jatropha curcas] |
| 18 |
Hb_000213_120 |
0.1404189834 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 19 |
Hb_002836_100 |
0.1411445388 |
- |
- |
PREDICTED: nudix hydrolase 3 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000059_020 |
0.141700149 |
- |
- |
PREDICTED: F-box protein SKP2A [Jatropha curcas] |