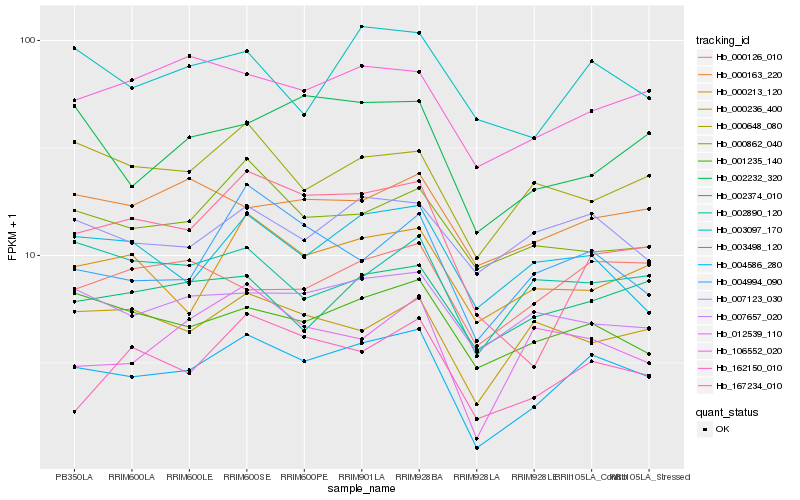
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004586_280 |
0.0 |
- |
- |
PREDICTED: GEM-like protein 1 [Jatropha curcas] |
| 2 |
Hb_167234_010 |
0.1237148853 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 13-like [Jatropha curcas] |
| 3 |
Hb_004994_090 |
0.1287291144 |
- |
- |
PREDICTED: uncharacterized protein LOC105643033 isoform X2 [Jatropha curcas] |
| 4 |
Hb_001235_140 |
0.129890031 |
- |
- |
PREDICTED: protein arginine N-methyltransferase 1.6 [Jatropha curcas] |
| 5 |
Hb_162150_010 |
0.1342449728 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000213_120 |
0.135147169 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 7 |
Hb_003097_170 |
0.1352557829 |
- |
- |
PREDICTED: protein EARLY RESPONSIVE TO DEHYDRATION 15-like isoform X3 [Jatropha curcas] |
| 8 |
Hb_106552_020 |
0.1367540001 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor RS2Z33 isoform X1 [Jatropha curcas] |
| 9 |
Hb_003498_120 |
0.1375135295 |
- |
- |
hypothetical protein POPTR_0015s12430g, partial [Populus trichocarpa] |
| 10 |
Hb_012539_110 |
0.1377445869 |
- |
- |
PREDICTED: ER membrane protein complex subunit 1 [Jatropha curcas] |
| 11 |
Hb_002232_320 |
0.1380558908 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 12 |
Hb_000236_400 |
0.1385128119 |
- |
- |
hypothetical protein JCGZ_01058 [Jatropha curcas] |
| 13 |
Hb_000648_080 |
0.1394851456 |
- |
- |
PREDICTED: serine/arginine repetitive matrix protein 1 isoform X2 [Populus euphratica] |
| 14 |
Hb_007657_020 |
0.13989133 |
- |
- |
PREDICTED: PRA1 family protein H isoform X1 [Jatropha curcas] |
| 15 |
Hb_002374_010 |
0.1402566906 |
- |
- |
PREDICTED: protein translocase subunit SECA2, chloroplastic [Jatropha curcas] |
| 16 |
Hb_007123_030 |
0.1403470835 |
- |
- |
PREDICTED: uncharacterized protein LOC105634056 isoform X3 [Jatropha curcas] |
| 17 |
Hb_000862_040 |
0.1411681452 |
- |
- |
ara4-interacting protein, putative [Ricinus communis] |
| 18 |
Hb_000126_010 |
0.141500433 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase RFK1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002890_120 |
0.1425016872 |
- |
- |
Nucleic acid binding,RNA binding isoform 1 [Theobroma cacao] |
| 20 |
Hb_000163_220 |
0.1446439355 |
- |
- |
hypothetical protein CISIN_1g022301mg [Citrus sinensis] |