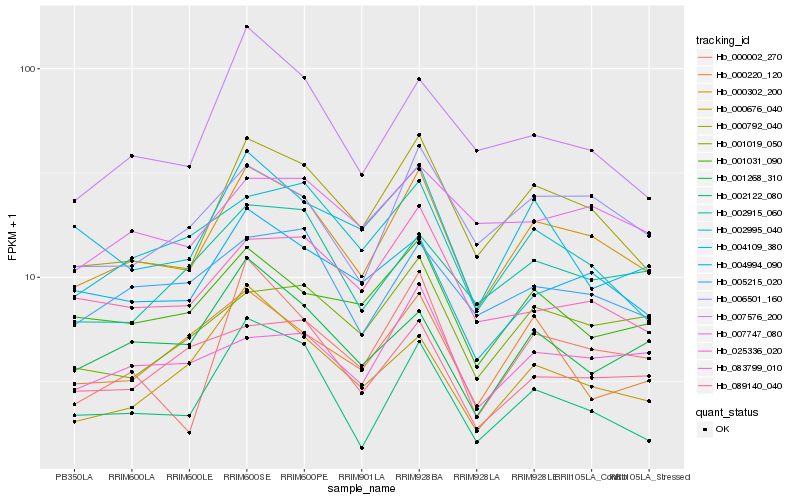
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000302_200 |
0.0 |
- |
- |
ornithine cyclodeaminase, putative [Ricinus communis] |
| 2 |
Hb_000792_040 |
0.0672751436 |
- |
- |
PREDICTED: steroid 5-alpha-reductase DET2 [Jatropha curcas] |
| 3 |
Hb_004994_090 |
0.0971083999 |
- |
- |
PREDICTED: uncharacterized protein LOC105643033 isoform X2 [Jatropha curcas] |
| 4 |
Hb_006501_160 |
0.0975193828 |
- |
- |
PREDICTED: hexokinase-1-like [Jatropha curcas] |
| 5 |
Hb_001031_090 |
0.1053278091 |
- |
- |
PREDICTED: translational activator GCN1 [Jatropha curcas] |
| 6 |
Hb_005215_020 |
0.1086574806 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB44 [Jatropha curcas] |
| 7 |
Hb_002995_040 |
0.1104003702 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase dyrk1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001019_050 |
0.1123434213 |
- |
- |
PREDICTED: importin-13 [Jatropha curcas] |
| 9 |
Hb_007576_200 |
0.1133794609 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 10 |
Hb_002915_060 |
0.1156083445 |
- |
- |
PREDICTED: serine/threonine-protein kinase CTR1-like [Jatropha curcas] |
| 11 |
Hb_083799_010 |
0.1158295473 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 12 |
Hb_002122_080 |
0.1178798981 |
- |
- |
- |
| 13 |
Hb_000676_040 |
0.118016012 |
- |
- |
hypothetical protein POPTR_0007s13320g [Populus trichocarpa] |
| 14 |
Hb_000002_270 |
0.1185334818 |
- |
- |
hypothetical protein VITISV_043897 [Vitis vinifera] |
| 15 |
Hb_089140_040 |
0.1194350236 |
- |
- |
PREDICTED: uncharacterized protein LOC105637966 [Jatropha curcas] |
| 16 |
Hb_025336_020 |
0.1196087455 |
- |
- |
PREDICTED: PP2A regulatory subunit TAP46 [Jatropha curcas] |
| 17 |
Hb_004109_380 |
0.1197373822 |
- |
- |
PREDICTED: callose synthase 9 isoform X4 [Pyrus x bretschneideri] |
| 18 |
Hb_001268_310 |
0.1200917431 |
- |
- |
- |
| 19 |
Hb_007747_080 |
0.1216625097 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000220_120 |
0.1221562491 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 isoform X2 [Jatropha curcas] |