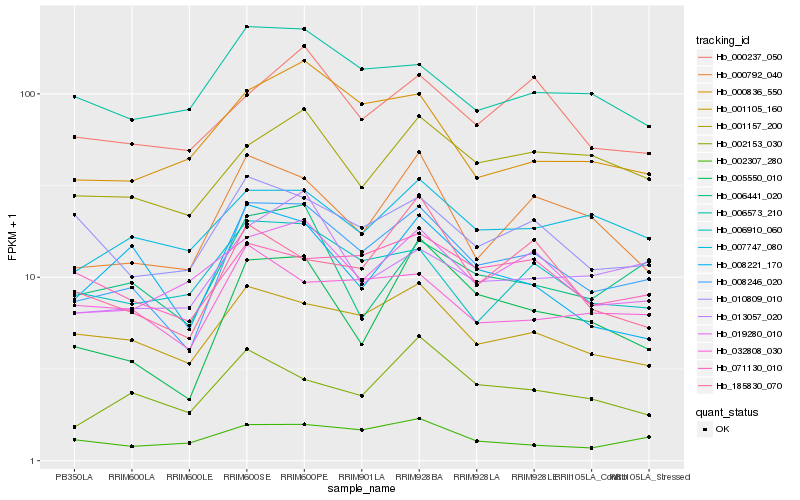
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008246_020 |
0.0 |
- |
- |
amidophosphoribosyltransferase, putative [Ricinus communis] |
| 2 |
Hb_001105_160 |
0.110927151 |
- |
- |
PREDICTED: protein phosphatase 2C 32 [Jatropha curcas] |
| 3 |
Hb_005550_010 |
0.1150394266 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 4-like [Jatropha curcas] |
| 4 |
Hb_000792_040 |
0.1179047829 |
- |
- |
PREDICTED: steroid 5-alpha-reductase DET2 [Jatropha curcas] |
| 5 |
Hb_019280_010 |
0.1235233427 |
- |
- |
PREDICTED: F-box protein At5g39450 isoform X2 [Jatropha curcas] |
| 6 |
Hb_006441_020 |
0.1270580328 |
- |
- |
PREDICTED: cadmium/zinc-transporting ATPase HMA3-like isoform X2 [Populus euphratica] |
| 7 |
Hb_001157_200 |
0.1273524288 |
- |
- |
PREDICTED: uncharacterized protein LOC105645131 [Jatropha curcas] |
| 8 |
Hb_006910_060 |
0.1284307268 |
- |
- |
PREDICTED: MACPF domain-containing protein At1g14780-like [Jatropha curcas] |
| 9 |
Hb_006573_210 |
0.1316325648 |
- |
- |
PREDICTED: protein SGT1 homolog [Jatropha curcas] |
| 10 |
Hb_010809_010 |
0.1319133508 |
- |
- |
Na+/H+ antiporter [Populus euphratica] |
| 11 |
Hb_007747_080 |
0.1322394992 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000237_050 |
0.1328135424 |
- |
- |
CP2 [Hevea brasiliensis] |
| 13 |
Hb_071130_010 |
0.1337068357 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000836_550 |
0.1341285295 |
- |
- |
PREDICTED: reticulon-like protein B5 [Jatropha curcas] |
| 15 |
Hb_008221_170 |
0.1341448184 |
transcription factor |
TF Family: BES1 |
PREDICTED: BES1/BZR1 homolog protein 4 isoform X1 [Populus euphratica] |
| 16 |
Hb_032808_030 |
0.1341904882 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_002153_030 |
0.1345923502 |
- |
- |
E3 ubiquitin-protein ligase UPL5 [Theobroma cacao] |
| 18 |
Hb_013057_020 |
0.1352508691 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RLIM-like [Jatropha curcas] |
| 19 |
Hb_002307_280 |
0.135888404 |
- |
- |
Endonuclease/exonuclease/phosphatase family protein isoform 3 [Theobroma cacao] |
| 20 |
Hb_185830_070 |
0.1359845637 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4-like isoform X1 [Jatropha curcas] |