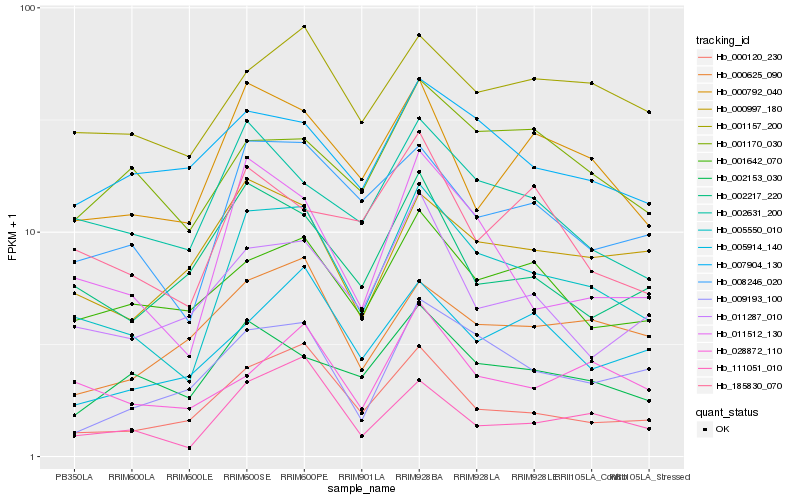
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005550_010 |
0.0 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 4-like [Jatropha curcas] |
| 2 |
Hb_008246_020 |
0.1150394266 |
- |
- |
amidophosphoribosyltransferase, putative [Ricinus communis] |
| 3 |
Hb_011512_130 |
0.1151718924 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At2g33170 [Jatropha curcas] |
| 4 |
Hb_000120_230 |
0.1282645749 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g33990-like [Jatropha curcas] |
| 5 |
Hb_000792_040 |
0.1334401882 |
- |
- |
PREDICTED: steroid 5-alpha-reductase DET2 [Jatropha curcas] |
| 6 |
Hb_111051_010 |
0.1347159261 |
- |
- |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 7 |
Hb_002631_200 |
0.141596963 |
- |
- |
PREDICTED: uncharacterized protein LOC105646171 isoform X2 [Jatropha curcas] |
| 8 |
Hb_001157_200 |
0.143932199 |
- |
- |
PREDICTED: uncharacterized protein LOC105645131 [Jatropha curcas] |
| 9 |
Hb_002153_030 |
0.1458354876 |
- |
- |
E3 ubiquitin-protein ligase UPL5 [Theobroma cacao] |
| 10 |
Hb_011287_010 |
0.146588723 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 11 |
Hb_001642_070 |
0.1468728333 |
- |
- |
PREDICTED: uncharacterized protein LOC105646431 [Jatropha curcas] |
| 12 |
Hb_009193_100 |
0.1496632449 |
- |
- |
hypothetical protein JCGZ_15709 [Jatropha curcas] |
| 13 |
Hb_028872_110 |
0.1499317076 |
- |
- |
PREDICTED: uncharacterized protein LOC105644776 [Jatropha curcas] |
| 14 |
Hb_185830_070 |
0.1505246737 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_000997_180 |
0.1518198629 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_005914_140 |
0.1521114149 |
- |
- |
PREDICTED: uncharacterized protein LOC105632183 [Jatropha curcas] |
| 17 |
Hb_002217_220 |
0.1546920577 |
- |
- |
hypothetical protein POPTR_0010s15930g [Populus trichocarpa] |
| 18 |
Hb_001170_030 |
0.1547579934 |
- |
- |
synaptonemal complex protein, putative [Ricinus communis] |
| 19 |
Hb_007904_130 |
0.1575352494 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 20 |
Hb_000625_090 |
0.1584932351 |
- |
- |
conserved hypothetical protein [Ricinus communis] |