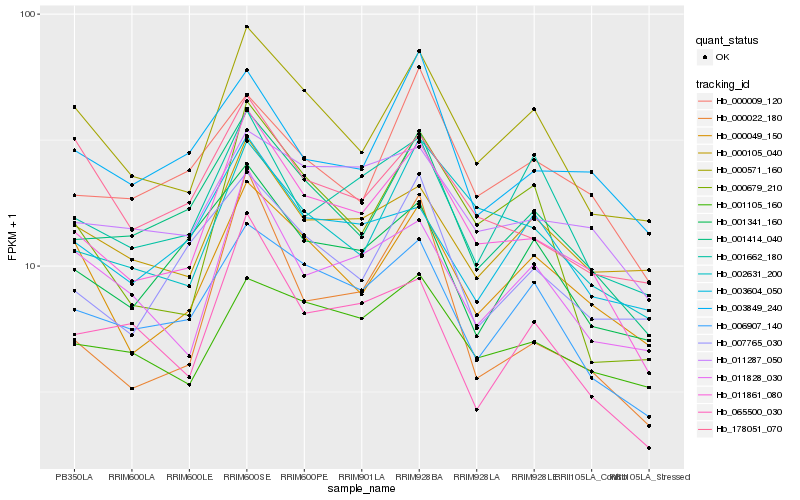
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011861_080 |
0.0 |
- |
- |
hypothetical protein JCGZ_06088 [Jatropha curcas] |
| 2 |
Hb_001414_040 |
0.0943401221 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 1-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_000571_160 |
0.0960626849 |
- |
- |
PREDICTED: ubiquitin-activating enzyme E1 1-like [Jatropha curcas] |
| 4 |
Hb_002631_200 |
0.101585883 |
- |
- |
PREDICTED: uncharacterized protein LOC105646171 isoform X2 [Jatropha curcas] |
| 5 |
Hb_006907_140 |
0.1038707839 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000049_150 |
0.1102229747 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 7 |
Hb_011287_050 |
0.1111698218 |
- |
- |
PREDICTED: uncharacterized protein LOC105644786 [Jatropha curcas] |
| 8 |
Hb_011828_030 |
0.1144602626 |
- |
- |
ubiquitin protein ligase E3a, putative [Ricinus communis] |
| 9 |
Hb_000679_210 |
0.1187030516 |
- |
- |
grr1, plant, putative [Ricinus communis] |
| 10 |
Hb_065500_030 |
0.1211716734 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI7 [Jatropha curcas] |
| 11 |
Hb_000009_120 |
0.123273512 |
- |
- |
PREDICTED: probable Xaa-Pro aminopeptidase P isoform X1 [Jatropha curcas] |
| 12 |
Hb_001105_160 |
0.1236374075 |
- |
- |
PREDICTED: protein phosphatase 2C 32 [Jatropha curcas] |
| 13 |
Hb_000022_180 |
0.1254484558 |
- |
- |
PREDICTED: RING finger and CHY zinc finger domain-containing protein 1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_178051_070 |
0.1260871891 |
- |
- |
PREDICTED: exocyst complex component EXO70B1-like [Jatropha curcas] |
| 15 |
Hb_001662_180 |
0.1282561754 |
- |
- |
PREDICTED: DNA-directed RNA polymerase II subunit 1-like isoform X5 [Citrus sinensis] |
| 16 |
Hb_000105_040 |
0.1283576435 |
- |
- |
PREDICTED: serine/threonine-protein kinase TOR isoform X2 [Jatropha curcas] |
| 17 |
Hb_001341_160 |
0.1285135712 |
- |
- |
PREDICTED: E3 SUMO-protein ligase SIZ1 isoform X2 [Jatropha curcas] |
| 18 |
Hb_003849_240 |
0.1285726634 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 19 |
Hb_003604_050 |
0.1291382194 |
- |
- |
PREDICTED: MAP3K epsilon protein kinase 1-like isoform X2 [Jatropha curcas] |
| 20 |
Hb_007765_030 |
0.1294829027 |
- |
- |
PREDICTED: neutral ceramidase [Jatropha curcas] |