| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
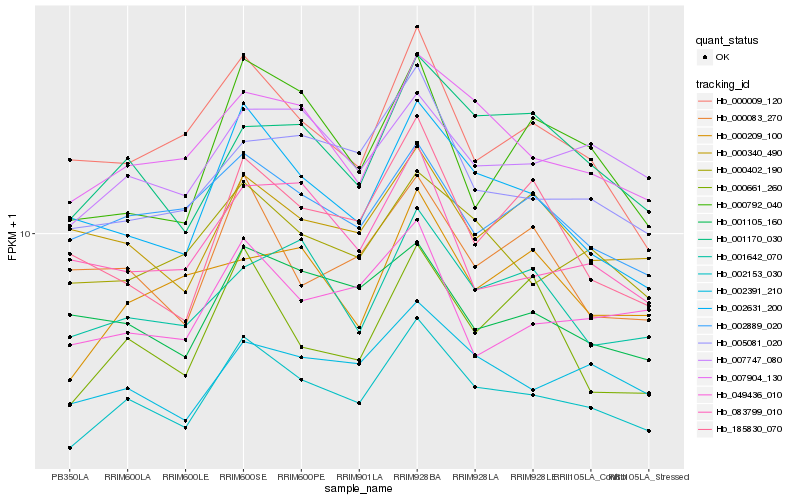
Hb_002153_030 |
0.0 |
- |
- |
E3 ubiquitin-protein ligase UPL5 [Theobroma cacao] |
| 2 |
Hb_002631_200 |
0.0962096227 |
- |
- |
PREDICTED: uncharacterized protein LOC105646171 isoform X2 [Jatropha curcas] |
| 3 |
Hb_007904_130 |
0.0971055173 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 4 |
Hb_002391_210 |
0.1047151336 |
- |
- |
PREDICTED: putative F-box protein At1g65770 [Jatropha curcas] |
| 5 |
Hb_001170_030 |
0.1089513655 |
- |
- |
synaptonemal complex protein, putative [Ricinus communis] |
| 6 |
Hb_000402_190 |
0.1091640015 |
- |
- |
PREDICTED: F-box protein At1g70590 [Jatropha curcas] |
| 7 |
Hb_005081_020 |
0.1121110251 |
- |
- |
PREDICTED: ninja-family protein mc410 [Jatropha curcas] |
| 8 |
Hb_000009_120 |
0.1128310299 |
- |
- |
PREDICTED: probable Xaa-Pro aminopeptidase P isoform X1 [Jatropha curcas] |
| 9 |
Hb_007747_080 |
0.1161378064 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000792_040 |
0.1183421327 |
- |
- |
PREDICTED: steroid 5-alpha-reductase DET2 [Jatropha curcas] |
| 11 |
Hb_000661_260 |
0.1199399027 |
- |
- |
PREDICTED: uncharacterized protein LOC105648304 [Jatropha curcas] |
| 12 |
Hb_002889_020 |
0.121756518 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 3 [Jatropha curcas] |
| 13 |
Hb_001642_070 |
0.1237575497 |
- |
- |
PREDICTED: uncharacterized protein LOC105646431 [Jatropha curcas] |
| 14 |
Hb_001105_160 |
0.1243271464 |
- |
- |
PREDICTED: protein phosphatase 2C 32 [Jatropha curcas] |
| 15 |
Hb_185830_070 |
0.1244442534 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_049436_010 |
0.125311377 |
- |
- |
PREDICTED: UPF0202 protein At1g10490-like [Glycine max] |
| 17 |
Hb_000083_270 |
0.1256429146 |
- |
- |
PREDICTED: putative uncharacterized protein At4g01020, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000340_490 |
0.1280090657 |
- |
- |
PREDICTED: ankyrin-1-like [Jatropha curcas] |
| 19 |
Hb_000209_100 |
0.1298061447 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 2 isoform X2 [Jatropha curcas] |
| 20 |
Hb_083799_010 |
0.1308824257 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |