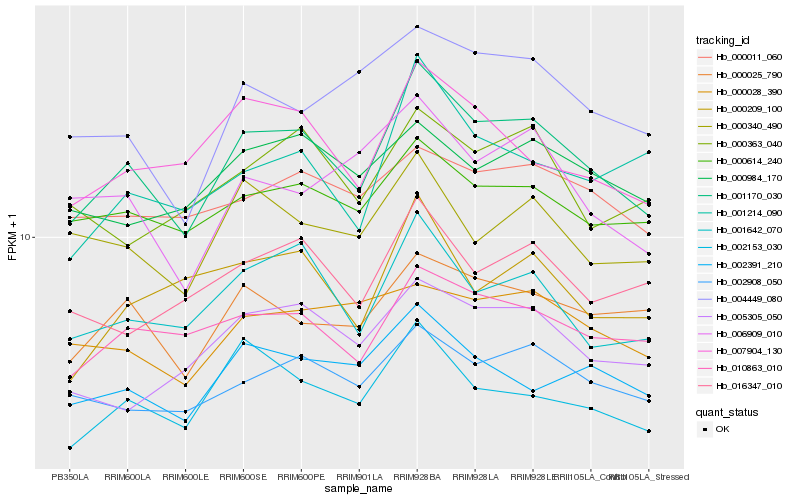
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001170_030 |
0.0 |
- |
- |
synaptonemal complex protein, putative [Ricinus communis] |
| 2 |
Hb_001642_070 |
0.0919574188 |
- |
- |
PREDICTED: uncharacterized protein LOC105646431 [Jatropha curcas] |
| 3 |
Hb_002908_050 |
0.0931549552 |
- |
- |
hypothetical protein CISIN_1g0022902mg, partial [Citrus sinensis] |
| 4 |
Hb_010863_010 |
0.0933568415 |
transcription factor |
TF Family: C3H |
nucleic acid binding protein, putative [Ricinus communis] |
| 5 |
Hb_007904_130 |
0.1006572166 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 6 |
Hb_000025_790 |
0.1007256975 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 3 [Jatropha curcas] |
| 7 |
Hb_000614_240 |
0.1026395831 |
- |
- |
PREDICTED: signal recognition particle subunit SRP72 [Jatropha curcas] |
| 8 |
Hb_002391_210 |
0.1057851652 |
- |
- |
PREDICTED: putative F-box protein At1g65770 [Jatropha curcas] |
| 9 |
Hb_002153_030 |
0.1089513655 |
- |
- |
E3 ubiquitin-protein ligase UPL5 [Theobroma cacao] |
| 10 |
Hb_016347_010 |
0.1107667962 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 11 |
Hb_004449_080 |
0.1111642343 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH3-like [Jatropha curcas] |
| 12 |
Hb_000340_490 |
0.1115809165 |
- |
- |
PREDICTED: ankyrin-1-like [Jatropha curcas] |
| 13 |
Hb_005305_050 |
0.1125587479 |
- |
- |
PREDICTED: armadillo repeat-containing protein 8 [Jatropha curcas] |
| 14 |
Hb_001214_090 |
0.1140379739 |
- |
- |
PREDICTED: acyl-coenzyme A oxidase 4, peroxisomal [Jatropha curcas] |
| 15 |
Hb_006909_010 |
0.1141944605 |
- |
- |
5-oxoprolinase, putative [Ricinus communis] |
| 16 |
Hb_000984_170 |
0.1155581972 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 17 |
Hb_000011_060 |
0.1167565646 |
- |
- |
PREDICTED: exportin-7 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000363_040 |
0.1175553381 |
- |
- |
Protein transport protein Sec24A, putative [Ricinus communis] |
| 19 |
Hb_000028_390 |
0.11757016 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 3-like [Jatropha curcas] |
| 20 |
Hb_000209_100 |
0.1178272679 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 2 isoform X2 [Jatropha curcas] |