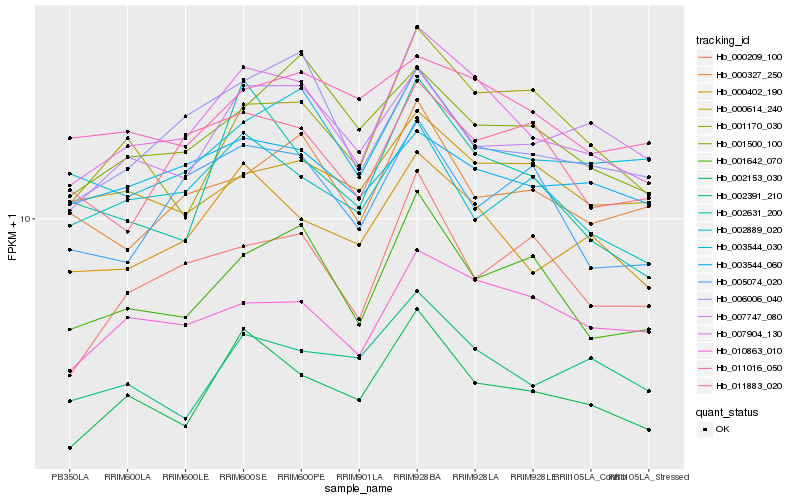
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007904_130 |
0.0 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 2 |
Hb_000402_190 |
0.0721611765 |
- |
- |
PREDICTED: F-box protein At1g70590 [Jatropha curcas] |
| 3 |
Hb_010863_010 |
0.0883679076 |
transcription factor |
TF Family: C3H |
nucleic acid binding protein, putative [Ricinus communis] |
| 4 |
Hb_001642_070 |
0.0909320872 |
- |
- |
PREDICTED: uncharacterized protein LOC105646431 [Jatropha curcas] |
| 5 |
Hb_003544_030 |
0.096222517 |
- |
- |
PREDICTED: citrate synthase, glyoxysomal [Jatropha curcas] |
| 6 |
Hb_002153_030 |
0.0971055173 |
- |
- |
E3 ubiquitin-protein ligase UPL5 [Theobroma cacao] |
| 7 |
Hb_001170_030 |
0.1006572166 |
- |
- |
synaptonemal complex protein, putative [Ricinus communis] |
| 8 |
Hb_011883_020 |
0.1024143643 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: aspartate--tRNA ligase, cytoplasmic-like [Jatropha curcas] |
| 9 |
Hb_007747_080 |
0.1028337908 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001500_100 |
0.1031738486 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002391_210 |
0.1034348163 |
- |
- |
PREDICTED: putative F-box protein At1g65770 [Jatropha curcas] |
| 12 |
Hb_011016_050 |
0.1037891717 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI7 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000327_250 |
0.1057920317 |
- |
- |
PREDICTED: macrophage erythroblast attacher [Jatropha curcas] |
| 14 |
Hb_006006_040 |
0.1060687696 |
- |
- |
S-methyl-5-thioribose kinase isoform 2 [Theobroma cacao] |
| 15 |
Hb_003544_060 |
0.1060807863 |
- |
- |
hypothetical protein JCGZ_22100 [Jatropha curcas] |
| 16 |
Hb_002889_020 |
0.1066633995 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 3 [Jatropha curcas] |
| 17 |
Hb_002631_200 |
0.1069849736 |
- |
- |
PREDICTED: uncharacterized protein LOC105646171 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000614_240 |
0.1073620252 |
- |
- |
PREDICTED: signal recognition particle subunit SRP72 [Jatropha curcas] |
| 19 |
Hb_005074_020 |
0.110727307 |
- |
- |
PREDICTED: uncharacterized protein LOC105644455 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000209_100 |
0.1117148351 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 2 isoform X2 [Jatropha curcas] |