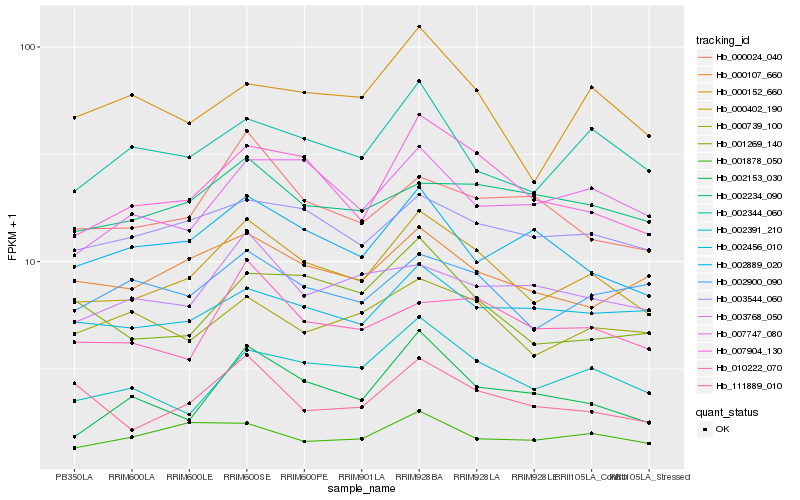
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000402_190 |
0.0 |
- |
- |
PREDICTED: F-box protein At1g70590 [Jatropha curcas] |
| 2 |
Hb_007904_130 |
0.0721611765 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 3 |
Hb_002391_210 |
0.0919914085 |
- |
- |
PREDICTED: putative F-box protein At1g65770 [Jatropha curcas] |
| 4 |
Hb_003544_060 |
0.0951724973 |
- |
- |
hypothetical protein JCGZ_22100 [Jatropha curcas] |
| 5 |
Hb_002344_060 |
0.0988564843 |
- |
- |
PREDICTED: methylthioribose-1-phosphate isomerase [Jatropha curcas] |
| 6 |
Hb_000107_660 |
0.0988906646 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 [Jatropha curcas] |
| 7 |
Hb_010222_070 |
0.1010236408 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_007747_080 |
0.101409524 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002456_010 |
0.1034337891 |
- |
- |
PREDICTED: protein kinase and PP2C-like domain-containing protein isoform X2 [Jatropha curcas] |
| 10 |
Hb_000739_100 |
0.1035069192 |
- |
- |
ku P80 DNA helicase, putative [Ricinus communis] |
| 11 |
Hb_002900_090 |
0.1036954571 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105646817 [Jatropha curcas] |
| 12 |
Hb_002889_020 |
0.1046847687 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 3 [Jatropha curcas] |
| 13 |
Hb_000152_660 |
0.1058357235 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 7 [Jatropha curcas] |
| 14 |
Hb_001269_140 |
0.1058680408 |
- |
- |
PREDICTED: nuclear pore complex protein NUP96 [Jatropha curcas] |
| 15 |
Hb_111889_010 |
0.105968188 |
- |
- |
PREDICTED: DNA ligase 4 [Jatropha curcas] |
| 16 |
Hb_002234_090 |
0.106098725 |
- |
- |
PREDICTED: probable transcriptional regulator SLK2 [Jatropha curcas] |
| 17 |
Hb_000024_040 |
0.1063586518 |
- |
- |
PREDICTED: ALG-2 interacting protein X [Jatropha curcas] |
| 18 |
Hb_003768_050 |
0.1086205656 |
- |
- |
PREDICTED: uncharacterized protein LOC105644152 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001878_050 |
0.1086280468 |
- |
- |
PREDICTED: uncharacterized protein LOC105646110 isoform X3 [Jatropha curcas] |
| 20 |
Hb_002153_030 |
0.1091640015 |
- |
- |
E3 ubiquitin-protein ligase UPL5 [Theobroma cacao] |