| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
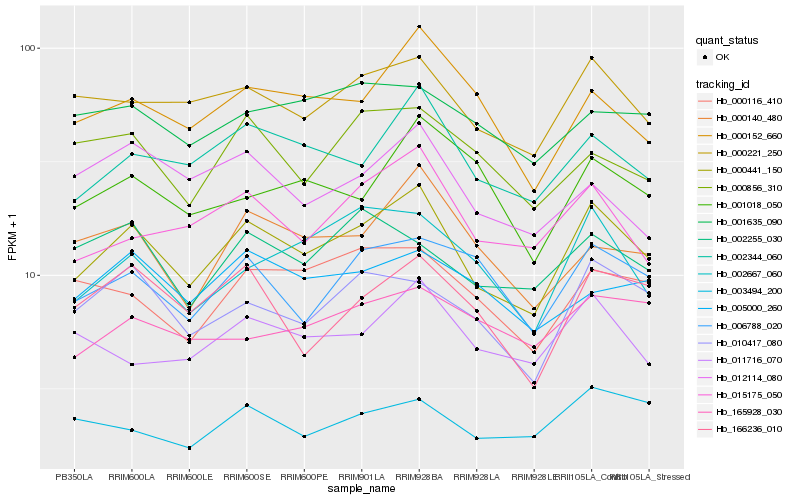
Hb_000441_150 |
0.0 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, root-type isozyme, chloroplastic isoform X2 [Jatropha curcas] |
| 2 |
Hb_002344_060 |
0.0893417253 |
- |
- |
PREDICTED: methylthioribose-1-phosphate isomerase [Jatropha curcas] |
| 3 |
Hb_000221_250 |
0.0906579794 |
- |
- |
hypothetical protein JCGZ_15814 [Jatropha curcas] |
| 4 |
Hb_015175_050 |
0.1005763695 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 10 isoform X3 [Jatropha curcas] |
| 5 |
Hb_010417_080 |
0.1014004764 |
- |
- |
PREDICTED: uncharacterized protein LOC105645742 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000152_660 |
0.1043493047 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 7 [Jatropha curcas] |
| 7 |
Hb_002667_060 |
0.1075196823 |
- |
- |
mannose-1-phosphate guanyltransferase, putative [Ricinus communis] |
| 8 |
Hb_006788_020 |
0.1078421628 |
- |
- |
hypothetical protein JCGZ_21864 [Jatropha curcas] |
| 9 |
Hb_166236_010 |
0.1082927069 |
- |
- |
- |
| 10 |
Hb_000140_480 |
0.1087930211 |
- |
- |
PREDICTED: uncharacterized protein LOC105637350 [Jatropha curcas] |
| 11 |
Hb_011716_070 |
0.1090869975 |
transcription factor |
TF Family: HRT |
PREDICTED: uncharacterized protein LOC105637858 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000116_410 |
0.1098401015 |
- |
- |
hypothetical protein JCGZ_22416 [Jatropha curcas] |
| 13 |
Hb_003494_200 |
0.1168479934 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 15-like [Jatropha curcas] |
| 14 |
Hb_005000_260 |
0.1176295581 |
- |
- |
recA family protein [Populus trichocarpa] |
| 15 |
Hb_000856_310 |
0.1179655595 |
- |
- |
PREDICTED: hypersensitive-induced response protein 1 [Jatropha curcas] |
| 16 |
Hb_001018_050 |
0.1190909774 |
- |
- |
PREDICTED: aspartate aminotransferase, cytoplasmic [Jatropha curcas] |
| 17 |
Hb_002255_030 |
0.1200049472 |
- |
- |
PREDICTED: INO80 complex subunit D-like [Jatropha curcas] |
| 18 |
Hb_165928_030 |
0.1203774166 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001635_090 |
0.1205515262 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 7 isoform X1 [Populus euphratica] |
| 20 |
Hb_012114_080 |
0.1209226885 |
- |
- |
gamma-glutamylcysteine synthetase [Hevea brasiliensis] |