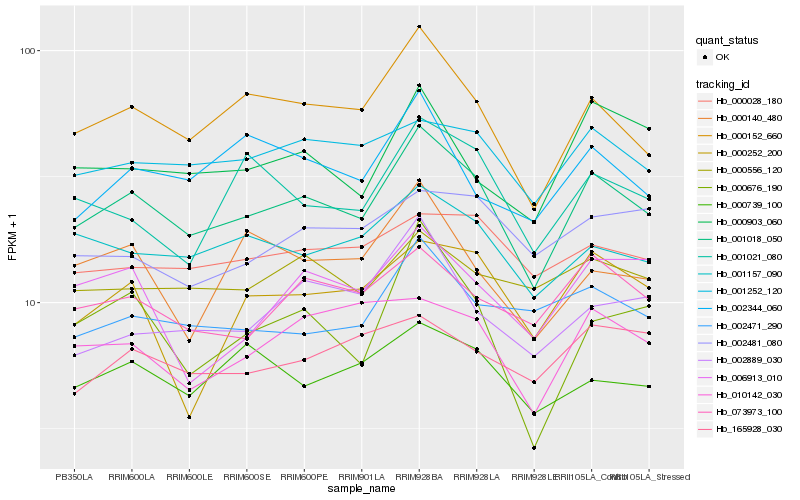
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001018_050 |
0.0 |
- |
- |
PREDICTED: aspartate aminotransferase, cytoplasmic [Jatropha curcas] |
| 2 |
Hb_000152_660 |
0.0641327175 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 7 [Jatropha curcas] |
| 3 |
Hb_001252_120 |
0.0880878548 |
- |
- |
PREDICTED: delta-1-pyrroline-5-carboxylate synthase-like [Gossypium raimondii] |
| 4 |
Hb_001157_090 |
0.0895467738 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase SINAT2-like [Jatropha curcas] |
| 5 |
Hb_073973_100 |
0.0900818151 |
- |
- |
PREDICTED: uncharacterized protein LOC105638910 [Jatropha curcas] |
| 6 |
Hb_000903_060 |
0.0967785788 |
- |
- |
PREDICTED: ras-related protein RABA5e [Jatropha curcas] |
| 7 |
Hb_010142_030 |
0.0979777159 |
- |
- |
PREDICTED: isoamylase 3, chloroplastic isoform X2 [Jatropha curcas] |
| 8 |
Hb_000739_100 |
0.0991695789 |
- |
- |
ku P80 DNA helicase, putative [Ricinus communis] |
| 9 |
Hb_000028_180 |
0.0993901015 |
- |
- |
PREDICTED: probable serine/threonine protein kinase IREH1 isoform X2 [Jatropha curcas] |
| 10 |
Hb_001021_080 |
0.0998075452 |
- |
- |
PREDICTED: autophagy-related protein 18a [Jatropha curcas] |
| 11 |
Hb_002471_290 |
0.1000257501 |
- |
- |
PREDICTED: vacuolar fusion protein CCZ1 homolog isoform X1 [Jatropha curcas] |
| 12 |
Hb_000556_120 |
0.1011354645 |
- |
- |
Spastin, putative [Ricinus communis] |
| 13 |
Hb_002344_060 |
0.1012853539 |
- |
- |
PREDICTED: methylthioribose-1-phosphate isomerase [Jatropha curcas] |
| 14 |
Hb_000676_190 |
0.1030579924 |
- |
- |
hypothetical protein JCGZ_08279 [Jatropha curcas] |
| 15 |
Hb_002889_030 |
0.1036195036 |
- |
- |
PREDICTED: uncharacterized protein At3g49140 [Jatropha curcas] |
| 16 |
Hb_000140_480 |
0.1048846556 |
- |
- |
PREDICTED: uncharacterized protein LOC105637350 [Jatropha curcas] |
| 17 |
Hb_165928_030 |
0.1051278261 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_006913_010 |
0.1083605848 |
- |
- |
PREDICTED: uncharacterized protein LOC105649144 [Jatropha curcas] |
| 19 |
Hb_002481_080 |
0.1094597543 |
- |
- |
PREDICTED: bifunctional 3-dehydroquinate dehydratase/shikimate dehydrogenase, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_000252_200 |
0.1102193655 |
- |
- |
conserved hypothetical protein [Ricinus communis] |