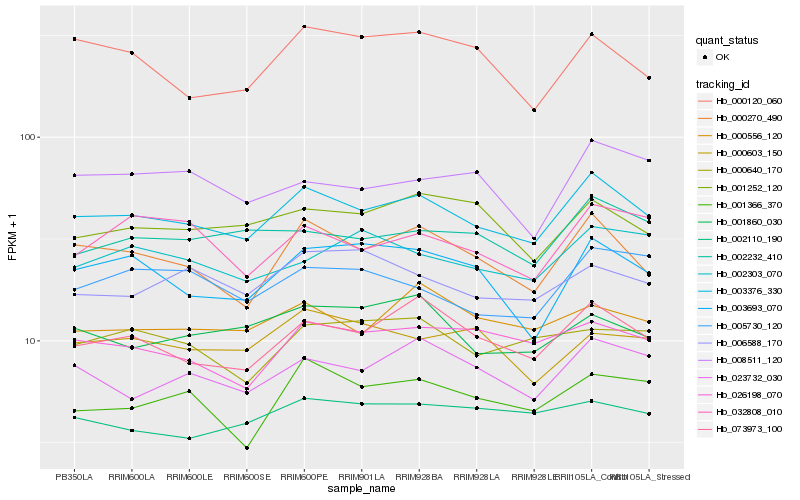
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003376_330 |
0.0 |
- |
- |
PREDICTED: gamma carbonic anhydrase 1, mitochondrial [Eucalyptus grandis] |
| 2 |
Hb_000270_490 |
0.0537542245 |
- |
- |
betaine aldehyde dehydrogenase 1 [Jatropha curcas] |
| 3 |
Hb_073973_100 |
0.0579765901 |
- |
- |
PREDICTED: uncharacterized protein LOC105638910 [Jatropha curcas] |
| 4 |
Hb_023732_030 |
0.0694612115 |
- |
- |
PREDICTED: proteinaceous RNase P 2-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_002232_410 |
0.0731067312 |
- |
- |
PREDICTED: L-galactose dehydrogenase [Jatropha curcas] |
| 6 |
Hb_026198_070 |
0.0742718114 |
- |
- |
PREDICTED: uncharacterized membrane protein At4g09580 [Jatropha curcas] |
| 7 |
Hb_003693_070 |
0.0751531475 |
- |
- |
PREDICTED: S-(hydroxymethyl)glutathione dehydrogenase [Jatropha curcas] |
| 8 |
Hb_000556_120 |
0.0761158368 |
- |
- |
Spastin, putative [Ricinus communis] |
| 9 |
Hb_001252_120 |
0.0761585338 |
- |
- |
PREDICTED: delta-1-pyrroline-5-carboxylate synthase-like [Gossypium raimondii] |
| 10 |
Hb_001860_030 |
0.0801040127 |
- |
- |
PREDICTED: cleavage stimulation factor subunit 77 isoform X5 [Jatropha curcas] |
| 11 |
Hb_001366_370 |
0.0810953449 |
- |
- |
PREDICTED: aarF domain-containing protein kinase 4 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000120_060 |
0.0812503524 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP20-1 [Populus euphratica] |
| 13 |
Hb_005730_120 |
0.0813545699 |
- |
- |
PREDICTED: protein WHI4 [Jatropha curcas] |
| 14 |
Hb_006588_170 |
0.0820873045 |
- |
- |
PREDICTED: OTU domain-containing protein At3g57810 [Jatropha curcas] |
| 15 |
Hb_002110_190 |
0.0822199168 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g27460 [Jatropha curcas] |
| 16 |
Hb_032808_010 |
0.0828471231 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin of 34 kDa [Jatropha curcas] |
| 17 |
Hb_002303_070 |
0.0829783359 |
- |
- |
PREDICTED: uncharacterized RNA-binding protein C1827.05c [Jatropha curcas] |
| 18 |
Hb_000640_170 |
0.0834445049 |
- |
- |
PREDICTED: uncharacterized protein LOC105642632 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000603_150 |
0.0839922644 |
- |
- |
PREDICTED: guanine nucleotide-binding protein alpha-1 subunit [Jatropha curcas] |
| 20 |
Hb_008511_120 |
0.0849720814 |
- |
- |
NADH-ubiquinone oxidoreductase 1, chain, putative [Ricinus communis] |