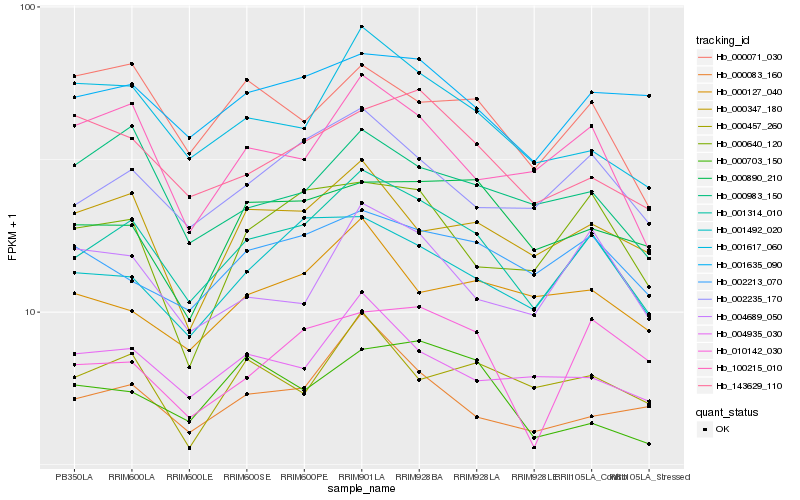
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001314_010 |
0.0 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein RICESLEEPER 1-like [Jatropha curcas] |
| 2 |
Hb_002235_170 |
0.0693814173 |
- |
- |
hypothetical protein CISIN_1g010208mg [Citrus sinensis] |
| 3 |
Hb_001492_020 |
0.0764635489 |
- |
- |
PREDICTED: diacylglycerol kinase 3-like [Jatropha curcas] |
| 4 |
Hb_001617_060 |
0.0767053486 |
- |
- |
PREDICTED: polyadenylate-binding protein RBP45B isoform X1 [Jatropha curcas] |
| 5 |
Hb_000083_160 |
0.0818585073 |
- |
- |
Cell division cycle protein, putative [Ricinus communis] |
| 6 |
Hb_100215_010 |
0.0825007871 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein R isoform X1 [Jatropha curcas] |
| 7 |
Hb_001635_090 |
0.082782895 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 7 isoform X1 [Populus euphratica] |
| 8 |
Hb_004935_030 |
0.0828277216 |
- |
- |
PREDICTED: uncharacterized protein LOC102628125 [Citrus sinensis] |
| 9 |
Hb_002213_070 |
0.0843629555 |
- |
- |
epsilon-adaptin family protein [Populus trichocarpa] |
| 10 |
Hb_143629_110 |
0.0859334637 |
- |
- |
glutamyl-tRNA synthetase, cytoplasmic, putative [Ricinus communis] |
| 11 |
Hb_000347_180 |
0.0868039153 |
- |
- |
PREDICTED: exocyst complex component EXO70A1-like [Jatropha curcas] |
| 12 |
Hb_000983_150 |
0.0871676973 |
- |
- |
PREDICTED: bifunctional nuclease 1 [Jatropha curcas] |
| 13 |
Hb_000703_150 |
0.0879389061 |
- |
- |
PREDICTED: F-box/LRR-repeat MAX2 homolog A [Jatropha curcas] |
| 14 |
Hb_000890_210 |
0.0894419494 |
- |
- |
PREDICTED: serine/threonine-protein kinase PBS1 isoform X1 [Eucalyptus grandis] |
| 15 |
Hb_000127_040 |
0.0900483283 |
- |
- |
PREDICTED: protein S-acyltransferase 8-like [Jatropha curcas] |
| 16 |
Hb_000071_030 |
0.0907497166 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase XBAT31 [Jatropha curcas] |
| 17 |
Hb_000457_260 |
0.0914200568 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g15690-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_004689_050 |
0.0915612497 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000640_120 |
0.0920277932 |
- |
- |
PREDICTED: uncharacterized protein LOC105642625 [Jatropha curcas] |
| 20 |
Hb_010142_030 |
0.0928676976 |
- |
- |
PREDICTED: isoamylase 3, chloroplastic isoform X2 [Jatropha curcas] |