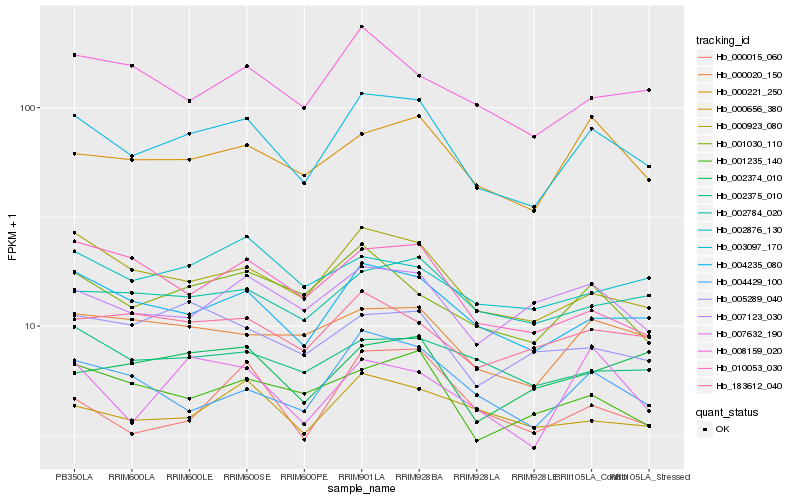
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003097_170 |
0.0 |
- |
- |
PREDICTED: protein EARLY RESPONSIVE TO DEHYDRATION 15-like isoform X3 [Jatropha curcas] |
| 2 |
Hb_004235_080 |
0.0660250117 |
- |
- |
PREDICTED: coiled-coil domain-containing protein SCD2 [Jatropha curcas] |
| 3 |
Hb_000923_080 |
0.0720466957 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 2A [Jatropha curcas] |
| 4 |
Hb_000221_250 |
0.0801143239 |
- |
- |
hypothetical protein JCGZ_15814 [Jatropha curcas] |
| 5 |
Hb_000020_150 |
0.0865672872 |
- |
- |
PREDICTED: uncharacterized protein LOC105636325 [Jatropha curcas] |
| 6 |
Hb_004429_100 |
0.0871590218 |
- |
- |
PREDICTED: asparagine synthetase domain-containing protein 1 [Jatropha curcas] |
| 7 |
Hb_001030_110 |
0.0877650499 |
- |
- |
PREDICTED: uncharacterized PKHD-type hydroxylase At1g22950 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001235_140 |
0.0908182278 |
- |
- |
PREDICTED: protein arginine N-methyltransferase 1.6 [Jatropha curcas] |
| 9 |
Hb_002784_020 |
0.0916765487 |
- |
- |
PREDICTED: probable protein arginine N-methyltransferase 3 [Jatropha curcas] |
| 10 |
Hb_007632_190 |
0.094041711 |
- |
- |
PREDICTED: inactive beta-amylase 4, chloroplastic isoform X5 [Camelina sativa] |
| 11 |
Hb_183612_040 |
0.0943232242 |
- |
- |
PREDICTED: uncharacterized protein LOC105632370 [Jatropha curcas] |
| 12 |
Hb_007123_030 |
0.0947717576 |
- |
- |
PREDICTED: uncharacterized protein LOC105634056 isoform X3 [Jatropha curcas] |
| 13 |
Hb_008159_020 |
0.095663828 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit beta-like protein [Jatropha curcas] |
| 14 |
Hb_000656_380 |
0.0959336876 |
- |
- |
PREDICTED: exportin-T [Jatropha curcas] |
| 15 |
Hb_000015_060 |
0.1008294959 |
- |
- |
PREDICTED: uncharacterized protein LOC104239844 isoform X2 [Nicotiana sylvestris] |
| 16 |
Hb_010053_030 |
0.1013055597 |
- |
- |
PREDICTED: tRNA (cytosine(34)-C(5))-methyltransferase [Jatropha curcas] |
| 17 |
Hb_002375_010 |
0.1014133221 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At1g03370 isoform X1 [Jatropha curcas] |
| 18 |
Hb_005289_040 |
0.1020177216 |
- |
- |
poly(A) polymerase, putative [Ricinus communis] |
| 19 |
Hb_002876_130 |
0.1023883305 |
- |
- |
PREDICTED: uncharacterized protein LOC105637143 [Jatropha curcas] |
| 20 |
Hb_002374_010 |
0.102614776 |
- |
- |
PREDICTED: protein translocase subunit SECA2, chloroplastic [Jatropha curcas] |