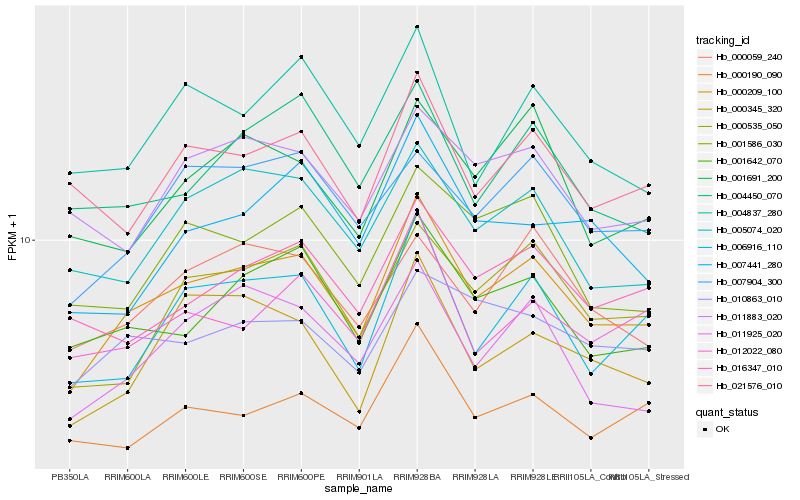
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000209_100 |
0.0 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 2 isoform X2 [Jatropha curcas] |
| 2 |
Hb_001642_070 |
0.0868954539 |
- |
- |
PREDICTED: uncharacterized protein LOC105646431 [Jatropha curcas] |
| 3 |
Hb_006916_110 |
0.0924957281 |
- |
- |
PREDICTED: probable tRNA (guanine(26)-N(2))-dimethyltransferase 2 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000535_050 |
0.0933623261 |
- |
- |
PREDICTED: cytosolic Fe-S cluster assembly factor narfl [Jatropha curcas] |
| 5 |
Hb_011925_020 |
0.0976518618 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 6 |
Hb_005074_020 |
0.0979759454 |
- |
- |
PREDICTED: uncharacterized protein LOC105644455 isoform X1 [Jatropha curcas] |
| 7 |
Hb_016347_010 |
0.0991045798 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 8 |
Hb_001586_030 |
0.0991914615 |
- |
- |
aspartyl-tRNA synthetase, putative [Ricinus communis] |
| 9 |
Hb_007904_300 |
0.0996290413 |
- |
- |
copine, putative [Ricinus communis] |
| 10 |
Hb_000345_320 |
0.1018799616 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein ALWAYS EARLY 2-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_000190_090 |
0.1042852066 |
- |
- |
PREDICTED: uncharacterized protein LOC105649930 [Jatropha curcas] |
| 12 |
Hb_000059_240 |
0.1044832597 |
- |
- |
PREDICTED: protein FRIGIDA [Jatropha curcas] |
| 13 |
Hb_010863_010 |
0.1046544469 |
transcription factor |
TF Family: C3H |
nucleic acid binding protein, putative [Ricinus communis] |
| 14 |
Hb_012022_080 |
0.1046842383 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 15 |
Hb_004837_280 |
0.1054462414 |
- |
- |
PREDICTED: uncharacterized protein LOC105648296 [Jatropha curcas] |
| 16 |
Hb_001691_200 |
0.1056828858 |
- |
- |
DNA-directed RNA polymerase II subunit RPB2 [Auxenochlorella protothecoides] |
| 17 |
Hb_021576_010 |
0.1064328607 |
- |
- |
PREDICTED: probable protein phosphatase 2C 60 [Jatropha curcas] |
| 18 |
Hb_004450_070 |
0.1073343283 |
- |
- |
PREDICTED: 2-hydroxyacyl-CoA lyase [Jatropha curcas] |
| 19 |
Hb_011883_020 |
0.1082715269 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: aspartate--tRNA ligase, cytoplasmic-like [Jatropha curcas] |
| 20 |
Hb_007441_280 |
0.1089951752 |
- |
- |
hypothetical protein JCGZ_14672 [Jatropha curcas] |