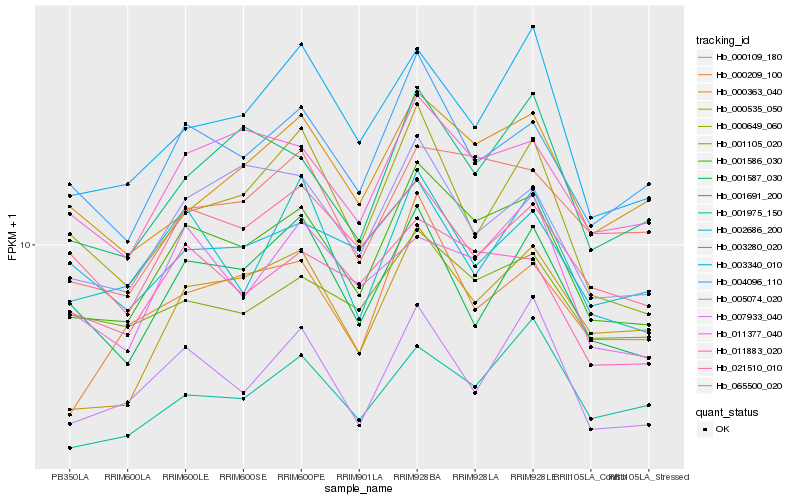
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001586_030 |
0.0 |
- |
- |
aspartyl-tRNA synthetase, putative [Ricinus communis] |
| 2 |
Hb_011883_020 |
0.0743658111 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: aspartate--tRNA ligase, cytoplasmic-like [Jatropha curcas] |
| 3 |
Hb_004096_110 |
0.0776512612 |
- |
- |
PREDICTED: T-complex protein 1 subunit gamma-like [Jatropha curcas] |
| 4 |
Hb_021510_010 |
0.0804485478 |
- |
- |
PREDICTED: deoxynucleoside triphosphate triphosphohydrolase SAMHD1 homolog [Jatropha curcas] |
| 5 |
Hb_005074_020 |
0.0829923445 |
- |
- |
PREDICTED: uncharacterized protein LOC105644455 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001691_200 |
0.0870372578 |
- |
- |
DNA-directed RNA polymerase II subunit RPB2 [Auxenochlorella protothecoides] |
| 7 |
Hb_000535_050 |
0.0878550181 |
- |
- |
PREDICTED: cytosolic Fe-S cluster assembly factor narfl [Jatropha curcas] |
| 8 |
Hb_000109_180 |
0.0882144384 |
- |
- |
PREDICTED: AP-1 complex subunit gamma-2-like [Jatropha curcas] |
| 9 |
Hb_007933_040 |
0.0890837375 |
- |
- |
PREDICTED: probable beta-D-xylosidase 6 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000363_040 |
0.0894442662 |
- |
- |
Protein transport protein Sec24A, putative [Ricinus communis] |
| 11 |
Hb_003340_010 |
0.0898098564 |
- |
- |
PREDICTED: ubiquitin-activating enzyme E1 1 [Vitis vinifera] |
| 12 |
Hb_001105_020 |
0.0928054627 |
- |
- |
expressed protein, putative [Ricinus communis] |
| 13 |
Hb_065500_020 |
0.0958530802 |
- |
- |
Exocyst complex component sec3A isoform 1 [Theobroma cacao] |
| 14 |
Hb_001587_030 |
0.0966762251 |
- |
- |
hypothetical protein JCGZ_09892 [Jatropha curcas] |
| 15 |
Hb_011377_040 |
0.0969430723 |
- |
- |
PREDICTED: uncharacterized protein LOC105647286 [Jatropha curcas] |
| 16 |
Hb_000649_060 |
0.0974951318 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase RIE1 [Jatropha curcas] |
| 17 |
Hb_001975_150 |
0.0984562982 |
- |
- |
PREDICTED: RINT1-like protein MAG2L [Jatropha curcas] |
| 18 |
Hb_002686_200 |
0.0989363861 |
- |
- |
glucosidase II beta subunit, putative [Ricinus communis] |
| 19 |
Hb_000209_100 |
0.0991914615 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 2 isoform X2 [Jatropha curcas] |
| 20 |
Hb_003280_020 |
0.0993628432 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4 isoform X1 [Jatropha curcas] |