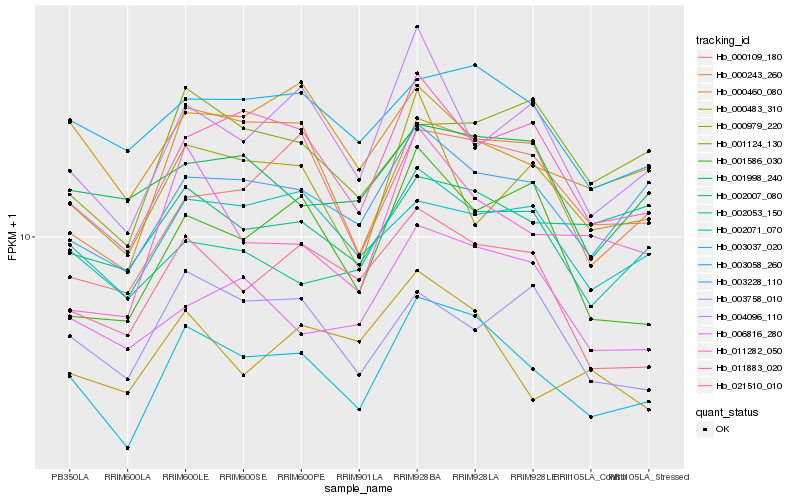
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003037_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_22910 [Jatropha curcas] |
| 2 |
Hb_011883_020 |
0.0848107478 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: aspartate--tRNA ligase, cytoplasmic-like [Jatropha curcas] |
| 3 |
Hb_003228_110 |
0.0864341093 |
- |
- |
PREDICTED: uncharacterized protein LOC105637245 [Jatropha curcas] |
| 4 |
Hb_003058_260 |
0.090417786 |
- |
- |
hypothetical protein POPTR_0003s13040g [Populus trichocarpa] |
| 5 |
Hb_000243_260 |
0.0921458841 |
- |
- |
PREDICTED: UPF0183 protein At3g51130 isoform X2 [Jatropha curcas] |
| 6 |
Hb_021510_010 |
0.092757001 |
- |
- |
PREDICTED: deoxynucleoside triphosphate triphosphohydrolase SAMHD1 homolog [Jatropha curcas] |
| 7 |
Hb_002071_070 |
0.0949583103 |
transcription factor |
TF Family: RB |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_006816_280 |
0.0999172782 |
- |
- |
PREDICTED: uncharacterized protein LOC105647448 isoform X2 [Jatropha curcas] |
| 9 |
Hb_004096_110 |
0.100941811 |
- |
- |
PREDICTED: T-complex protein 1 subunit gamma-like [Jatropha curcas] |
| 10 |
Hb_002053_150 |
0.1036395879 |
- |
- |
PREDICTED: small nuclear ribonucleoprotein-associated protein B' [Jatropha curcas] |
| 11 |
Hb_000460_080 |
0.1040102237 |
- |
- |
acyl-coenzyme A binding domain containing, putative [Ricinus communis] |
| 12 |
Hb_000483_310 |
0.1062569292 |
- |
- |
PREDICTED: WD-40 repeat-containing protein MSI4-like [Jatropha curcas] |
| 13 |
Hb_011282_050 |
0.1066220351 |
- |
- |
PREDICTED: diaminopimelate decarboxylase 2, chloroplastic-like [Jatropha curcas] |
| 14 |
Hb_002007_080 |
0.1074896469 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 4-like [Jatropha curcas] |
| 15 |
Hb_001124_130 |
0.1077397649 |
transcription factor |
TF Family: TUB |
phosphoric diester hydrolase, putative [Ricinus communis] |
| 16 |
Hb_000109_180 |
0.1077710094 |
- |
- |
PREDICTED: AP-1 complex subunit gamma-2-like [Jatropha curcas] |
| 17 |
Hb_001586_030 |
0.1085532782 |
- |
- |
aspartyl-tRNA synthetase, putative [Ricinus communis] |
| 18 |
Hb_003758_010 |
0.1085905032 |
transcription factor |
TF Family: VOZ |
PREDICTED: transcription factor VOZ1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001998_240 |
0.1103897925 |
- |
- |
PREDICTED: small nuclear ribonucleoprotein-associated protein B' [Jatropha curcas] |
| 20 |
Hb_000979_220 |
0.1109263526 |
- |
- |
PREDICTED: uncharacterized protein LOC105634085 isoform X1 [Jatropha curcas] |