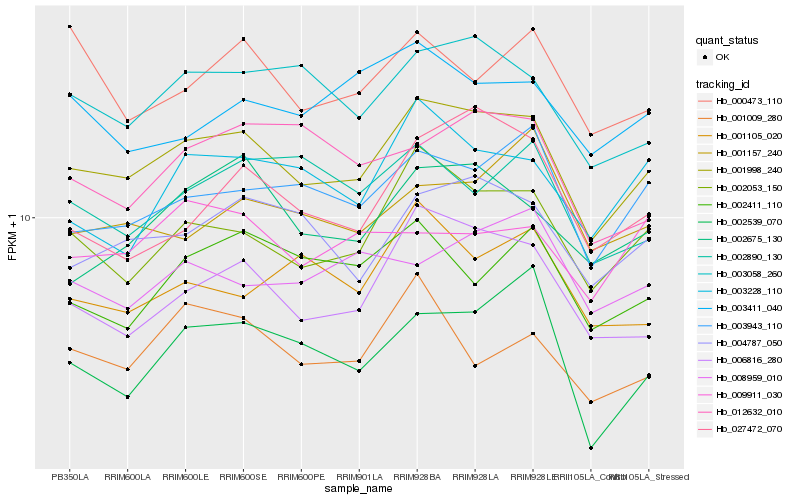
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001998_240 |
0.0 |
- |
- |
PREDICTED: small nuclear ribonucleoprotein-associated protein B' [Jatropha curcas] |
| 2 |
Hb_002053_150 |
0.0556229039 |
- |
- |
PREDICTED: small nuclear ribonucleoprotein-associated protein B' [Jatropha curcas] |
| 3 |
Hb_006816_280 |
0.0648353983 |
- |
- |
PREDICTED: uncharacterized protein LOC105647448 isoform X2 [Jatropha curcas] |
| 4 |
Hb_004787_050 |
0.0654696054 |
- |
- |
PREDICTED: RINT1-like protein MAG2 [Jatropha curcas] |
| 5 |
Hb_003058_260 |
0.066562022 |
- |
- |
hypothetical protein POPTR_0003s13040g [Populus trichocarpa] |
| 6 |
Hb_003943_110 |
0.0677526209 |
transcription factor |
TF Family: MYB-related |
Zuotin, putative [Ricinus communis] |
| 7 |
Hb_002675_130 |
0.0747859675 |
- |
- |
PREDICTED: N-alpha-acetyltransferase 16, NatA auxiliary subunit isoform X1 [Jatropha curcas] |
| 8 |
Hb_003228_110 |
0.0784543409 |
- |
- |
PREDICTED: uncharacterized protein LOC105637245 [Jatropha curcas] |
| 9 |
Hb_027472_070 |
0.0825790492 |
- |
- |
PREDICTED: uncharacterized protein LOC105646172 [Jatropha curcas] |
| 10 |
Hb_001157_240 |
0.0833340475 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 37-like [Jatropha curcas] |
| 11 |
Hb_003411_040 |
0.0840278988 |
- |
- |
unnamed protein product [Coffea canephora] |
| 12 |
Hb_002539_070 |
0.0841452643 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g26460, mitochondrial [Jatropha curcas] |
| 13 |
Hb_001009_280 |
0.0848259361 |
- |
- |
PREDICTED: telomere length regulation protein TEL2 homolog [Jatropha curcas] |
| 14 |
Hb_002411_110 |
0.0851579956 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit RPC2 [Jatropha curcas] |
| 15 |
Hb_009911_030 |
0.0853479295 |
transcription factor |
TF Family: FAR1 |
far-red impaired response 1 -like protein [Gossypium arboreum] |
| 16 |
Hb_002890_130 |
0.0854410151 |
- |
- |
tip120, putative [Ricinus communis] |
| 17 |
Hb_012632_010 |
0.0858230153 |
- |
- |
PREDICTED: uncharacterized protein LOC105640019 [Jatropha curcas] |
| 18 |
Hb_008959_010 |
0.0863580888 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000473_110 |
0.0865695066 |
- |
- |
PREDICTED: PP2A regulatory subunit TAP46 [Jatropha curcas] |
| 20 |
Hb_001105_020 |
0.0868053326 |
- |
- |
expressed protein, putative [Ricinus communis] |