| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
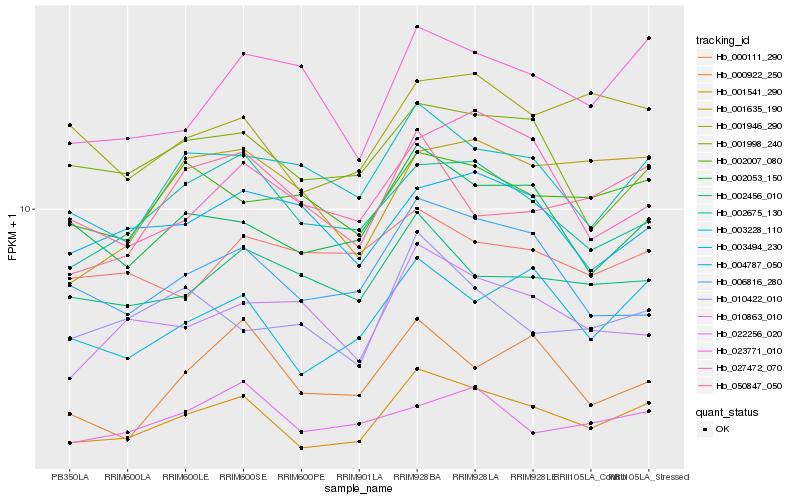
Hb_001541_290 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644977 [Jatropha curcas] |
| 2 |
Hb_003494_230 |
0.0681675668 |
- |
- |
starch synthase isoform II [Manihot esculenta] |
| 3 |
Hb_002053_150 |
0.0803676276 |
- |
- |
PREDICTED: small nuclear ribonucleoprotein-associated protein B' [Jatropha curcas] |
| 4 |
Hb_002675_130 |
0.0812316832 |
- |
- |
PREDICTED: N-alpha-acetyltransferase 16, NatA auxiliary subunit isoform X1 [Jatropha curcas] |
| 5 |
Hb_003228_110 |
0.0840172571 |
- |
- |
PREDICTED: uncharacterized protein LOC105637245 [Jatropha curcas] |
| 6 |
Hb_001946_290 |
0.0872960416 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 51-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_001998_240 |
0.0889836992 |
- |
- |
PREDICTED: small nuclear ribonucleoprotein-associated protein B' [Jatropha curcas] |
| 8 |
Hb_002456_010 |
0.089983385 |
- |
- |
PREDICTED: protein kinase and PP2C-like domain-containing protein isoform X2 [Jatropha curcas] |
| 9 |
Hb_006816_280 |
0.0912960808 |
- |
- |
PREDICTED: uncharacterized protein LOC105647448 isoform X2 [Jatropha curcas] |
| 10 |
Hb_002007_080 |
0.0924244458 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 4-like [Jatropha curcas] |
| 11 |
Hb_023771_010 |
0.0958376559 |
- |
- |
basic helix-loop-helix-containing protein, putative [Ricinus communis] |
| 12 |
Hb_022256_020 |
0.0959778927 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g35130, chloroplastic [Jatropha curcas] |
| 13 |
Hb_027472_070 |
0.098592253 |
- |
- |
PREDICTED: uncharacterized protein LOC105646172 [Jatropha curcas] |
| 14 |
Hb_010863_010 |
0.0986193548 |
transcription factor |
TF Family: C3H |
nucleic acid binding protein, putative [Ricinus communis] |
| 15 |
Hb_001635_190 |
0.0998479259 |
- |
- |
RNA m5u methyltransferase, putative [Ricinus communis] |
| 16 |
Hb_010422_010 |
0.1000031521 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |
| 17 |
Hb_004787_050 |
0.1007997916 |
- |
- |
PREDICTED: RINT1-like protein MAG2 [Jatropha curcas] |
| 18 |
Hb_050847_050 |
0.1011682227 |
- |
- |
PREDICTED: uncharacterized protein LOC105649140 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000922_250 |
0.1030674954 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 20 |
Hb_000111_290 |
0.1048945079 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase KEG [Jatropha curcas] |