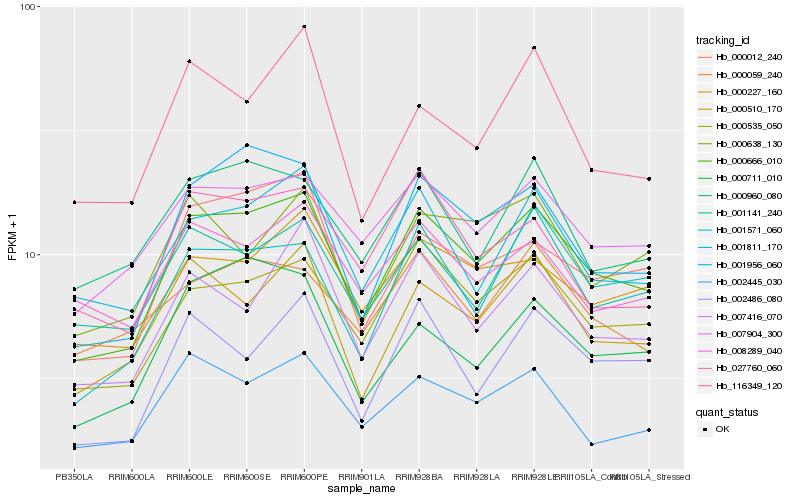
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000666_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000535_050 |
0.06966827 |
- |
- |
PREDICTED: cytosolic Fe-S cluster assembly factor narfl [Jatropha curcas] |
| 3 |
Hb_001956_060 |
0.0776488635 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 3C-like [Jatropha curcas] |
| 4 |
Hb_007904_300 |
0.079085961 |
- |
- |
copine, putative [Ricinus communis] |
| 5 |
Hb_000510_170 |
0.085790361 |
- |
- |
Glycosyltransferase QUASIMODO1, putative [Ricinus communis] |
| 6 |
Hb_001811_170 |
0.0863103 |
- |
- |
dynamin, putative [Ricinus communis] |
| 7 |
Hb_000012_240 |
0.0868351544 |
- |
- |
PREDICTED: ferrochelatase-2, chloroplastic [Jatropha curcas] |
| 8 |
Hb_002445_030 |
0.0896622484 |
- |
- |
hypothetical protein JCGZ_12656 [Jatropha curcas] |
| 9 |
Hb_001141_240 |
0.0909572065 |
- |
- |
PREDICTED: folate-biopterin transporter 1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000227_160 |
0.0925233264 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF1A [Jatropha curcas] |
| 11 |
Hb_001571_060 |
0.0928185579 |
- |
- |
PREDICTED: uncharacterized protein LOC105643976 [Jatropha curcas] |
| 12 |
Hb_007416_070 |
0.0935124155 |
transcription factor |
TF Family: B3 |
hypothetical protein JCGZ_07038 [Jatropha curcas] |
| 13 |
Hb_000960_080 |
0.0968457925 |
- |
- |
PREDICTED: topless-related protein 1-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_002486_080 |
0.1008780388 |
- |
- |
PREDICTED: cytosolic endo-beta-N-acetylglucosaminidase isoform X1 [Jatropha curcas] |
| 15 |
Hb_027760_060 |
0.1009940236 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 16 |
Hb_000711_010 |
0.1029073575 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 6B isoform X1 [Jatropha curcas] |
| 17 |
Hb_008289_040 |
0.1029966241 |
- |
- |
PREDICTED: suppressor of mec-8 and unc-52 protein homolog 1 [Jatropha curcas] |
| 18 |
Hb_000059_240 |
0.1030453798 |
- |
- |
PREDICTED: protein FRIGIDA [Jatropha curcas] |
| 19 |
Hb_000638_130 |
0.1030491858 |
- |
- |
PREDICTED: SAL1 phosphatase-like [Jatropha curcas] |
| 20 |
Hb_116349_120 |
0.1041100887 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit alpha-1, mitochondrial isoform X1 [Jatropha curcas] |