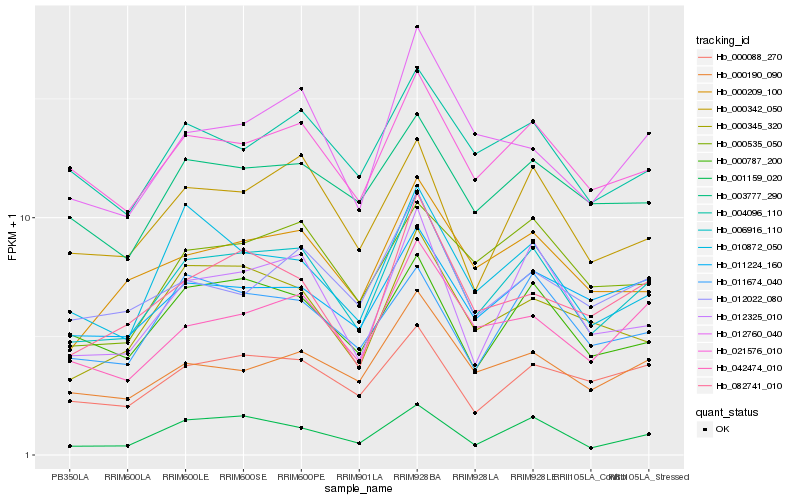
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006916_110 |
0.0 |
- |
- |
PREDICTED: probable tRNA (guanine(26)-N(2))-dimethyltransferase 2 isoform X2 [Jatropha curcas] |
| 2 |
Hb_012325_010 |
0.0811792549 |
- |
- |
hypothetical protein RCOM_0068670 [Ricinus communis] |
| 3 |
Hb_042474_010 |
0.0874954922 |
- |
- |
PREDICTED: adenylyltransferase and sulfurtransferase MOCS3 [Jatropha curcas] |
| 4 |
Hb_021576_010 |
0.0891611367 |
- |
- |
PREDICTED: probable protein phosphatase 2C 60 [Jatropha curcas] |
| 5 |
Hb_000342_050 |
0.0906960713 |
- |
- |
hypothetical protein POPTR_0006s24710g [Populus trichocarpa] |
| 6 |
Hb_000209_100 |
0.0924957281 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 2 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000190_090 |
0.0934878536 |
- |
- |
PREDICTED: uncharacterized protein LOC105649930 [Jatropha curcas] |
| 8 |
Hb_000787_200 |
0.0945935856 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 9 |
Hb_000088_270 |
0.0958244669 |
- |
- |
PREDICTED: uncharacterized protein LOC105637003 [Jatropha curcas] |
| 10 |
Hb_082741_010 |
0.0976364969 |
- |
- |
PREDICTED: uncharacterized protein LOC105649930 [Jatropha curcas] |
| 11 |
Hb_001159_020 |
0.098007899 |
- |
- |
- |
| 12 |
Hb_010872_050 |
0.099247955 |
- |
- |
hypothetical protein B456_007G078100 [Gossypium raimondii] |
| 13 |
Hb_012760_040 |
0.0993144174 |
- |
- |
PREDICTED: aconitate hydratase 1 [Jatropha curcas] |
| 14 |
Hb_004096_110 |
0.1007490313 |
- |
- |
PREDICTED: T-complex protein 1 subunit gamma-like [Jatropha curcas] |
| 15 |
Hb_000535_050 |
0.1015972331 |
- |
- |
PREDICTED: cytosolic Fe-S cluster assembly factor narfl [Jatropha curcas] |
| 16 |
Hb_000345_320 |
0.1022177377 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein ALWAYS EARLY 2-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_011674_040 |
0.1024570076 |
- |
- |
PREDICTED: uncharacterized protein LOC105648163 isoform X1 [Jatropha curcas] |
| 18 |
Hb_011224_160 |
0.10274512 |
transcription factor, rubber biosynthesis |
TF Family: HSF, Gene Name: Farnesyl diphosphate synthase |
PREDICTED: heat stress transcription factor A-5 [Jatropha curcas] |
| 19 |
Hb_012022_080 |
0.1032520202 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 20 |
Hb_003777_290 |
0.1032853805 |
- |
- |
polypyrimidine tract binding protein, putative [Ricinus communis] |