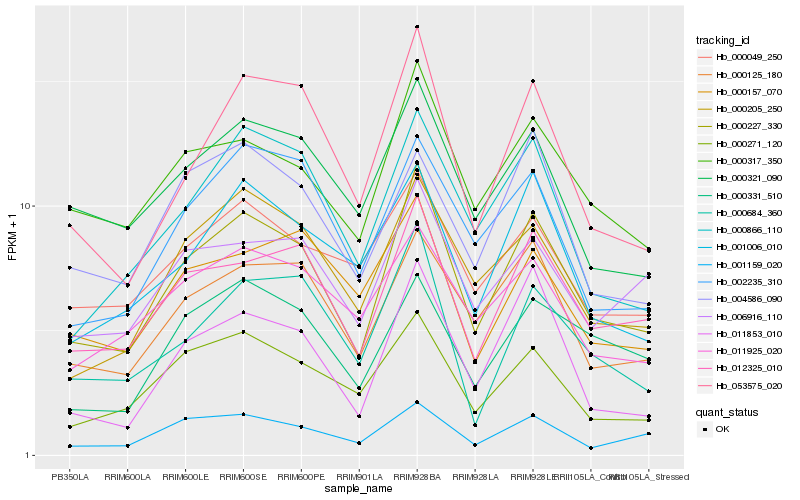
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000227_330 |
0.0 |
- |
- |
Acyl-CoA synthetase [Ricinus communis] |
| 2 |
Hb_012325_010 |
0.0817396345 |
- |
- |
hypothetical protein RCOM_0068670 [Ricinus communis] |
| 3 |
Hb_053575_020 |
0.0927010882 |
- |
- |
PREDICTED: RNA-binding protein 39 [Jatropha curcas] |
| 4 |
Hb_000125_180 |
0.0984357548 |
- |
- |
PREDICTED: flowering time control protein FPA isoform X1 [Jatropha curcas] |
| 5 |
Hb_000317_350 |
0.1016661785 |
- |
- |
PREDICTED: protein SUPPRESSOR OF GENE SILENCING 3 [Jatropha curcas] |
| 6 |
Hb_000205_250 |
0.1045172708 |
- |
- |
PREDICTED: uncharacterized protein LOC105647762 [Jatropha curcas] |
| 7 |
Hb_000271_120 |
0.1063194981 |
- |
- |
PREDICTED: U-box domain-containing protein 13-like [Nelumbo nucifera] |
| 8 |
Hb_000866_110 |
0.1094715724 |
- |
- |
PREDICTED: uncharacterized protein LOC105642980 isoform X2 [Jatropha curcas] |
| 9 |
Hb_001159_020 |
0.1123192018 |
- |
- |
- |
| 10 |
Hb_002235_310 |
0.1141998311 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 [Jatropha curcas] |
| 11 |
Hb_000321_090 |
0.1151788381 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, endoplasmic reticulum-type-like [Jatropha curcas] |
| 12 |
Hb_011853_010 |
0.115533009 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 [Jatropha curcas] |
| 13 |
Hb_006916_110 |
0.1168216086 |
- |
- |
PREDICTED: probable tRNA (guanine(26)-N(2))-dimethyltransferase 2 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000684_360 |
0.116863235 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_16454 [Jatropha curcas] |
| 15 |
Hb_001006_010 |
0.1169138017 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 16 |
Hb_000331_510 |
0.1189065588 |
- |
- |
PREDICTED: peroxisome biogenesis factor 10-like [Jatropha curcas] |
| 17 |
Hb_011925_020 |
0.1236642424 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 18 |
Hb_000049_250 |
0.1239997536 |
- |
- |
PREDICTED: uncharacterized protein LOC105644455 isoform X1 [Jatropha curcas] |
| 19 |
Hb_004586_090 |
0.1244786767 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 16 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000157_070 |
0.1261051817 |
- |
- |
PREDICTED: alpha-N-acetylglucosaminidase [Jatropha curcas] |