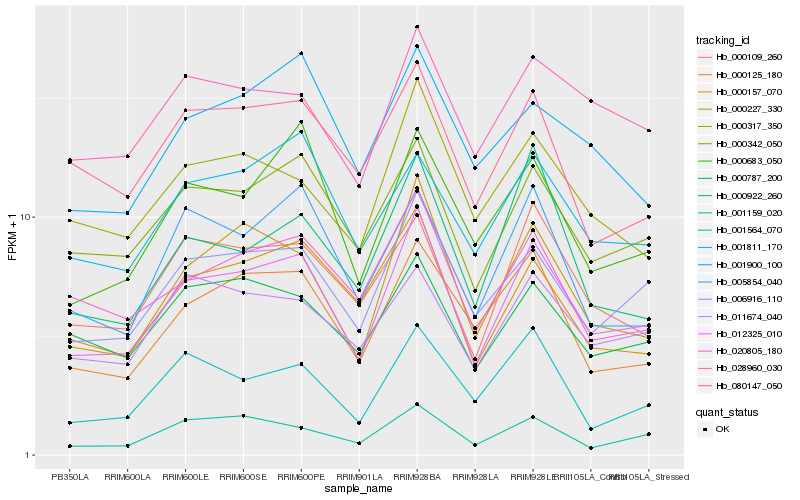
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012325_010 |
0.0 |
- |
- |
hypothetical protein RCOM_0068670 [Ricinus communis] |
| 2 |
Hb_006916_110 |
0.0811792549 |
- |
- |
PREDICTED: probable tRNA (guanine(26)-N(2))-dimethyltransferase 2 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000227_330 |
0.0817396345 |
- |
- |
Acyl-CoA synthetase [Ricinus communis] |
| 4 |
Hb_000342_050 |
0.0853624485 |
- |
- |
hypothetical protein POPTR_0006s24710g [Populus trichocarpa] |
| 5 |
Hb_000125_180 |
0.0980633476 |
- |
- |
PREDICTED: flowering time control protein FPA isoform X1 [Jatropha curcas] |
| 6 |
Hb_000157_070 |
0.0994190815 |
- |
- |
PREDICTED: alpha-N-acetylglucosaminidase [Jatropha curcas] |
| 7 |
Hb_020805_180 |
0.1008213582 |
- |
- |
PREDICTED: epidermal growth factor receptor substrate 15-like 1 [Jatropha curcas] |
| 8 |
Hb_000683_050 |
0.1031052975 |
- |
- |
PREDICTED: probable carbohydrate esterase At4g34215 [Jatropha curcas] |
| 9 |
Hb_000109_260 |
0.1040728658 |
- |
- |
PREDICTED: WAT1-related protein At3g28050-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_000787_200 |
0.1048903912 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_000317_350 |
0.1069017985 |
- |
- |
PREDICTED: protein SUPPRESSOR OF GENE SILENCING 3 [Jatropha curcas] |
| 12 |
Hb_001159_020 |
0.1093106664 |
- |
- |
- |
| 13 |
Hb_005854_040 |
0.1096898527 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_011674_040 |
0.1112171799 |
- |
- |
PREDICTED: uncharacterized protein LOC105648163 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001811_170 |
0.1118657656 |
- |
- |
dynamin, putative [Ricinus communis] |
| 16 |
Hb_001564_070 |
0.1125396363 |
- |
- |
PREDICTED: G patch domain-containing protein TGH [Jatropha curcas] |
| 17 |
Hb_028960_030 |
0.1127885891 |
- |
- |
Short-chain dehydrogenase-reductase B [Theobroma cacao] |
| 18 |
Hb_080147_050 |
0.1156447142 |
- |
- |
PREDICTED: probable plastidic glucose transporter 2 [Jatropha curcas] |
| 19 |
Hb_000922_260 |
0.1157462696 |
- |
- |
PREDICTED: pantothenate kinase 2 [Jatropha curcas] |
| 20 |
Hb_001900_100 |
0.116511651 |
- |
- |
PREDICTED: uncharacterized protein LOC105632664 isoform X1 [Jatropha curcas] |