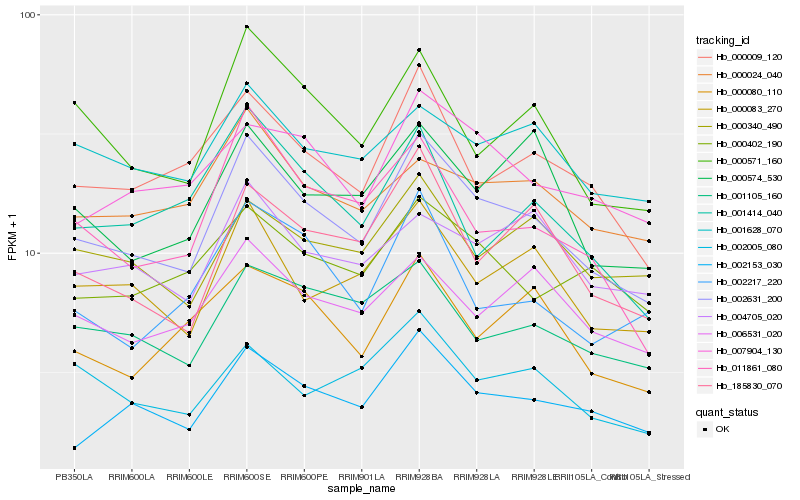
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002631_200 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105646171 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000083_270 |
0.0957310446 |
- |
- |
PREDICTED: putative uncharacterized protein At4g01020, chloroplastic [Jatropha curcas] |
| 3 |
Hb_002153_030 |
0.0962096227 |
- |
- |
E3 ubiquitin-protein ligase UPL5 [Theobroma cacao] |
| 4 |
Hb_000009_120 |
0.0969851923 |
- |
- |
PREDICTED: probable Xaa-Pro aminopeptidase P isoform X1 [Jatropha curcas] |
| 5 |
Hb_000571_160 |
0.0977727678 |
- |
- |
PREDICTED: ubiquitin-activating enzyme E1 1-like [Jatropha curcas] |
| 6 |
Hb_011861_080 |
0.101585883 |
- |
- |
hypothetical protein JCGZ_06088 [Jatropha curcas] |
| 7 |
Hb_185830_070 |
0.1042271041 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_007904_130 |
0.1069849736 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 9 |
Hb_001105_160 |
0.1084439883 |
- |
- |
PREDICTED: protein phosphatase 2C 32 [Jatropha curcas] |
| 10 |
Hb_006531_020 |
0.1102069615 |
- |
- |
PREDICTED: autophagy-related protein 13 [Jatropha curcas] |
| 11 |
Hb_000402_190 |
0.111236084 |
- |
- |
PREDICTED: F-box protein At1g70590 [Jatropha curcas] |
| 12 |
Hb_001628_070 |
0.1118758216 |
- |
- |
hect ubiquitin-protein ligase, putative [Ricinus communis] |
| 13 |
Hb_000024_040 |
0.1128186759 |
- |
- |
PREDICTED: ALG-2 interacting protein X [Jatropha curcas] |
| 14 |
Hb_000340_490 |
0.1147264567 |
- |
- |
PREDICTED: ankyrin-1-like [Jatropha curcas] |
| 15 |
Hb_000574_530 |
0.114808456 |
- |
- |
PREDICTED: pre-mRNA-processing-splicing factor 8 [Jatropha curcas] |
| 16 |
Hb_002217_220 |
0.1154385746 |
- |
- |
hypothetical protein POPTR_0010s15930g [Populus trichocarpa] |
| 17 |
Hb_002005_080 |
0.1155136614 |
- |
- |
PREDICTED: uncharacterized protein LOC105649750 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000080_110 |
0.1157537527 |
- |
- |
PREDICTED: GPI ethanolamine phosphate transferase 3 isoform X1 [Jatropha curcas] |
| 19 |
Hb_004705_020 |
0.1173915631 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 3 [Jatropha curcas] |
| 20 |
Hb_001414_040 |
0.118791641 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 1-like isoform X1 [Jatropha curcas] |