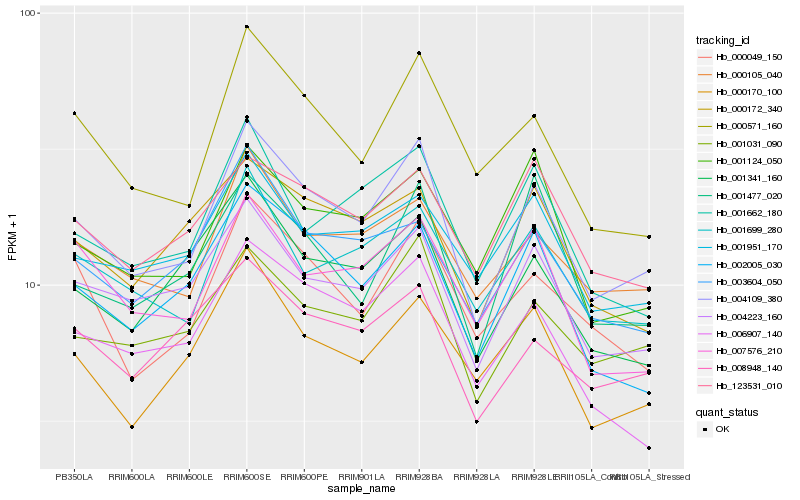
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004109_380 |
0.0 |
- |
- |
PREDICTED: callose synthase 9 isoform X4 [Pyrus x bretschneideri] |
| 2 |
Hb_001031_090 |
0.0734005062 |
- |
- |
PREDICTED: translational activator GCN1 [Jatropha curcas] |
| 3 |
Hb_004223_160 |
0.0752055688 |
- |
- |
PREDICTED: 5'-3' exoribonuclease 3 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000571_160 |
0.0850752873 |
- |
- |
PREDICTED: ubiquitin-activating enzyme E1 1-like [Jatropha curcas] |
| 5 |
Hb_000105_040 |
0.0858214062 |
- |
- |
PREDICTED: serine/threonine-protein kinase TOR isoform X2 [Jatropha curcas] |
| 6 |
Hb_002005_030 |
0.0858658256 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 7 |
Hb_001699_280 |
0.0889777939 |
- |
- |
PREDICTED: transformation/transcription domain-associated protein [Jatropha curcas] |
| 8 |
Hb_001341_160 |
0.09036687 |
- |
- |
PREDICTED: E3 SUMO-protein ligase SIZ1 isoform X2 [Jatropha curcas] |
| 9 |
Hb_001662_180 |
0.0906448568 |
- |
- |
PREDICTED: DNA-directed RNA polymerase II subunit 1-like isoform X5 [Citrus sinensis] |
| 10 |
Hb_000049_150 |
0.090756111 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 11 |
Hb_006907_140 |
0.0918643133 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000170_100 |
0.0919169855 |
transcription factor |
TF Family: C3H |
hypothetical protein JCGZ_12159 [Jatropha curcas] |
| 13 |
Hb_003604_050 |
0.0942007409 |
- |
- |
PREDICTED: MAP3K epsilon protein kinase 1-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_008948_140 |
0.0943843418 |
- |
- |
PREDICTED: nuclear pore complex protein NUP205 [Jatropha curcas] |
| 15 |
Hb_001477_020 |
0.095837267 |
transcription factor |
TF Family: HB |
PREDICTED: uncharacterized protein LOC105646886 isoform X3 [Jatropha curcas] |
| 16 |
Hb_001951_170 |
0.0960817559 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 17 |
Hb_000172_340 |
0.0971221836 |
- |
- |
PREDICTED: uncharacterized protein LOC101261327 [Solanum lycopersicum] |
| 18 |
Hb_001124_050 |
0.097125443 |
transcription factor |
TF Family: SNF2 |
PREDICTED: chromatin structure-remodeling complex protein SYD-like [Jatropha curcas] |
| 19 |
Hb_007576_210 |
0.0972601488 |
- |
- |
PREDICTED: protein furry homolog [Jatropha curcas] |
| 20 |
Hb_123531_010 |
0.0974281573 |
- |
- |
PREDICTED: nuclear pore complex protein NUP98A isoform X1 [Jatropha curcas] |