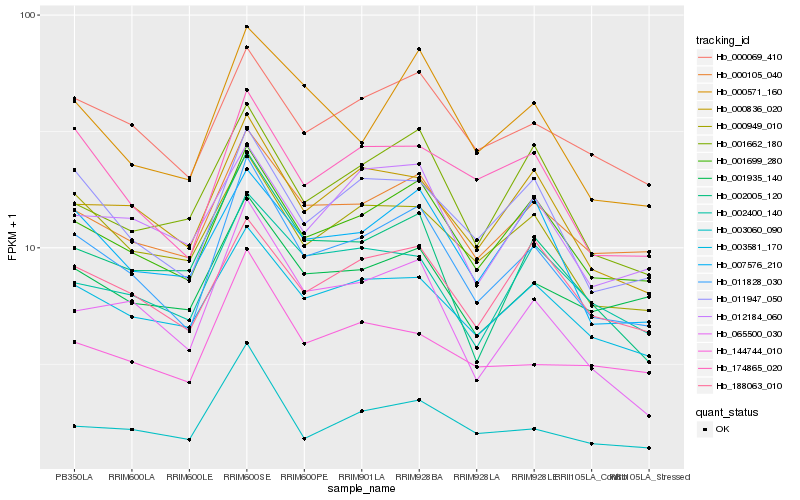
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011828_030 |
0.0 |
- |
- |
ubiquitin protein ligase E3a, putative [Ricinus communis] |
| 2 |
Hb_001699_280 |
0.0667412447 |
- |
- |
PREDICTED: transformation/transcription domain-associated protein [Jatropha curcas] |
| 3 |
Hb_000105_040 |
0.0775090133 |
- |
- |
PREDICTED: serine/threonine-protein kinase TOR isoform X2 [Jatropha curcas] |
| 4 |
Hb_011947_050 |
0.0779870107 |
- |
- |
E3 ubiquitin protein ligase upl2, putative [Ricinus communis] |
| 5 |
Hb_000069_410 |
0.0845696019 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 6 |
Hb_000836_020 |
0.0847144169 |
transcription factor |
TF Family: SNF2 |
Chromo domain protein, putative [Ricinus communis] |
| 7 |
Hb_003581_170 |
0.0849188685 |
- |
- |
PREDICTED: alpha-amylase 3, chloroplastic [Jatropha curcas] |
| 8 |
Hb_174865_020 |
0.0873762077 |
- |
- |
E3 ubiquitin protein ligase upl2, putative [Ricinus communis] |
| 9 |
Hb_000949_010 |
0.0893988059 |
- |
- |
PREDICTED: regulator of nonsense transcripts UPF2 [Jatropha curcas] |
| 10 |
Hb_000571_160 |
0.0913611955 |
- |
- |
PREDICTED: ubiquitin-activating enzyme E1 1-like [Jatropha curcas] |
| 11 |
Hb_007576_210 |
0.093518683 |
- |
- |
PREDICTED: protein furry homolog [Jatropha curcas] |
| 12 |
Hb_065500_030 |
0.0955008551 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI7 [Jatropha curcas] |
| 13 |
Hb_001935_140 |
0.0969638142 |
- |
- |
CCR4-NOT transcription complex subunit 1 [Gossypium arboreum] |
| 14 |
Hb_188063_010 |
0.0991970164 |
- |
- |
hypothetical protein RCOM_1469330 [Ricinus communis] |
| 15 |
Hb_001662_180 |
0.1003185206 |
- |
- |
PREDICTED: DNA-directed RNA polymerase II subunit 1-like isoform X5 [Citrus sinensis] |
| 16 |
Hb_003060_090 |
0.1025768784 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_20174 [Jatropha curcas] |
| 17 |
Hb_002400_140 |
0.103058709 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 18 |
Hb_012184_060 |
0.1042833446 |
- |
- |
PREDICTED: nuclear pore complex protein NUP1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002005_120 |
0.1049340752 |
- |
- |
PREDICTED: protein MEI2-like 5 [Jatropha curcas] |
| 20 |
Hb_144744_010 |
0.1059267458 |
- |
- |
hypothetical protein JCGZ_19531 [Jatropha curcas] |