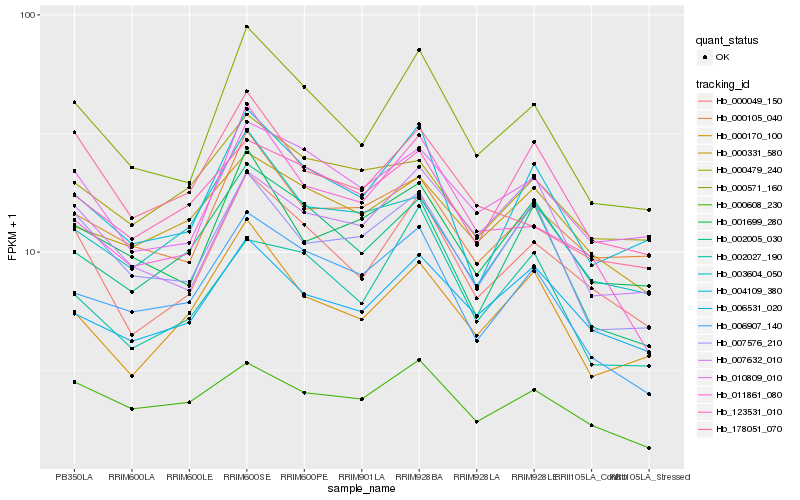
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000049_150 |
0.0 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 2 |
Hb_000571_160 |
0.0660760883 |
- |
- |
PREDICTED: ubiquitin-activating enzyme E1 1-like [Jatropha curcas] |
| 3 |
Hb_010809_010 |
0.0782003432 |
- |
- |
Na+/H+ antiporter [Populus euphratica] |
| 4 |
Hb_004109_380 |
0.090756111 |
- |
- |
PREDICTED: callose synthase 9 isoform X4 [Pyrus x bretschneideri] |
| 5 |
Hb_000170_100 |
0.0980281111 |
transcription factor |
TF Family: C3H |
hypothetical protein JCGZ_12159 [Jatropha curcas] |
| 6 |
Hb_000105_040 |
0.0998384634 |
- |
- |
PREDICTED: serine/threonine-protein kinase TOR isoform X2 [Jatropha curcas] |
| 7 |
Hb_006531_020 |
0.1004968336 |
- |
- |
PREDICTED: autophagy-related protein 13 [Jatropha curcas] |
| 8 |
Hb_007576_210 |
0.1093793893 |
- |
- |
PREDICTED: protein furry homolog [Jatropha curcas] |
| 9 |
Hb_002027_190 |
0.1095220582 |
- |
- |
PREDICTED: autophagy-related protein 18g [Jatropha curcas] |
| 10 |
Hb_001699_280 |
0.1099606703 |
- |
- |
PREDICTED: transformation/transcription domain-associated protein [Jatropha curcas] |
| 11 |
Hb_002005_030 |
0.110034769 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 12 |
Hb_011861_080 |
0.1102229747 |
- |
- |
hypothetical protein JCGZ_06088 [Jatropha curcas] |
| 13 |
Hb_000331_580 |
0.1103058624 |
- |
- |
PREDICTED: programmed cell death protein 4-like [Jatropha curcas] |
| 14 |
Hb_000608_230 |
0.1114012884 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 15 |
Hb_003604_050 |
0.1115866643 |
- |
- |
PREDICTED: MAP3K epsilon protein kinase 1-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_006907_140 |
0.1121781334 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_123531_010 |
0.1139128851 |
- |
- |
PREDICTED: nuclear pore complex protein NUP98A isoform X1 [Jatropha curcas] |
| 18 |
Hb_007632_010 |
0.1148308128 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000479_240 |
0.1149088514 |
- |
- |
PREDICTED: probable ubiquitin conjugation factor E4 isoform X2 [Jatropha curcas] |
| 20 |
Hb_178051_070 |
0.1162326155 |
- |
- |
PREDICTED: exocyst complex component EXO70B1-like [Jatropha curcas] |