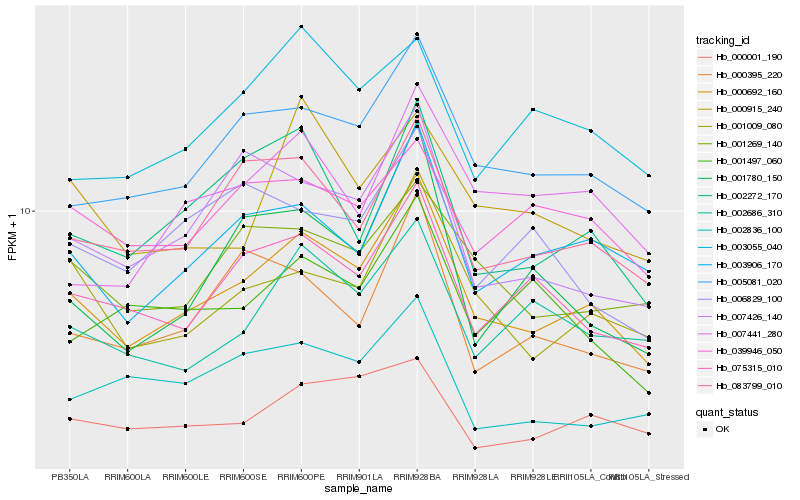
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000692_160 |
0.0 |
- |
- |
Transportin 1 isoform 1 [Theobroma cacao] |
| 2 |
Hb_005081_020 |
0.0832743128 |
- |
- |
PREDICTED: ninja-family protein mc410 [Jatropha curcas] |
| 3 |
Hb_003906_170 |
0.0921561765 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 4 |
Hb_083799_010 |
0.0959233629 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 5 |
Hb_001009_080 |
0.0963069335 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_075315_010 |
0.1001469068 |
- |
- |
ornithine aminotransferase [Camellia sinensis] |
| 7 |
Hb_002272_170 |
0.1042176389 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase-like protein 1 [Jatropha curcas] |
| 8 |
Hb_001269_140 |
0.1078756256 |
- |
- |
PREDICTED: nuclear pore complex protein NUP96 [Jatropha curcas] |
| 9 |
Hb_000001_190 |
0.1117776606 |
- |
- |
hypothetical protein CISIN_1g0082522mg, partial [Citrus sinensis] |
| 10 |
Hb_002686_310 |
0.1135411271 |
- |
- |
PREDICTED: aquaporin SIP1-1 [Jatropha curcas] |
| 11 |
Hb_007426_140 |
0.1137284996 |
- |
- |
ornithine aminotransferase, putative [Ricinus communis] |
| 12 |
Hb_000395_220 |
0.1172964875 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 13 |
Hb_039946_050 |
0.1219080218 |
- |
- |
catalytic, putative [Ricinus communis] |
| 14 |
Hb_001497_060 |
0.1222454065 |
- |
- |
PREDICTED: acyl-coenzyme A oxidase 3, peroxisomal-like [Jatropha curcas] |
| 15 |
Hb_001780_150 |
0.1233501131 |
- |
- |
ceramidase, putative [Ricinus communis] |
| 16 |
Hb_007441_280 |
0.1246215321 |
- |
- |
hypothetical protein JCGZ_14672 [Jatropha curcas] |
| 17 |
Hb_002836_100 |
0.1260896016 |
- |
- |
PREDICTED: nudix hydrolase 3 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000915_240 |
0.1269917129 |
- |
- |
catalytic, putative [Ricinus communis] |
| 19 |
Hb_003055_040 |
0.1285468073 |
- |
- |
PREDICTED: secretory carrier-associated membrane protein 3-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_006829_100 |
0.1285582327 |
- |
- |
conserved hypothetical protein [Ricinus communis] |