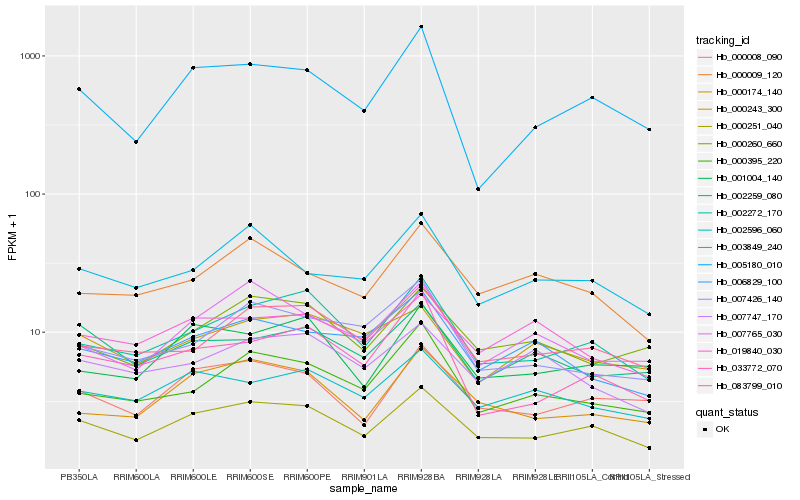
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000251_040 |
0.0 |
- |
- |
PREDICTED: DNA polymerase alpha catalytic subunit isoform X1 [Jatropha curcas] |
| 2 |
Hb_002272_170 |
0.0758835919 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase-like protein 1 [Jatropha curcas] |
| 3 |
Hb_000008_090 |
0.0859082347 |
- |
- |
PREDICTED: exosome complex component MTR3 [Jatropha curcas] |
| 4 |
Hb_000174_140 |
0.0961923132 |
- |
- |
JHL23J11.8 [Jatropha curcas] |
| 5 |
Hb_083799_010 |
0.0973058526 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 6 |
Hb_005180_010 |
0.1012475425 |
- |
- |
cyclophilin [Hevea brasiliensis] |
| 7 |
Hb_001004_140 |
0.1064864495 |
- |
- |
hypothetical protein JCGZ_26893 [Jatropha curcas] |
| 8 |
Hb_003849_240 |
0.1067577116 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 9 |
Hb_002596_060 |
0.1072479332 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000260_660 |
0.1084239577 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000395_220 |
0.1121252381 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 12 |
Hb_000009_120 |
0.1132917331 |
- |
- |
PREDICTED: probable Xaa-Pro aminopeptidase P isoform X1 [Jatropha curcas] |
| 13 |
Hb_007765_030 |
0.1157622798 |
- |
- |
PREDICTED: neutral ceramidase [Jatropha curcas] |
| 14 |
Hb_007426_140 |
0.1213058682 |
- |
- |
ornithine aminotransferase, putative [Ricinus communis] |
| 15 |
Hb_000243_300 |
0.1226158784 |
- |
- |
catalytic, putative [Ricinus communis] |
| 16 |
Hb_002259_080 |
0.1232415078 |
- |
- |
protein transport protein sec23, putative [Ricinus communis] |
| 17 |
Hb_006829_100 |
0.1235880972 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_033772_070 |
0.1250031852 |
- |
- |
PREDICTED: uncharacterized protein LOC105633681 [Jatropha curcas] |
| 19 |
Hb_007747_170 |
0.1258159807 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 [Jatropha curcas] |
| 20 |
Hb_019840_030 |
0.1263352351 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |