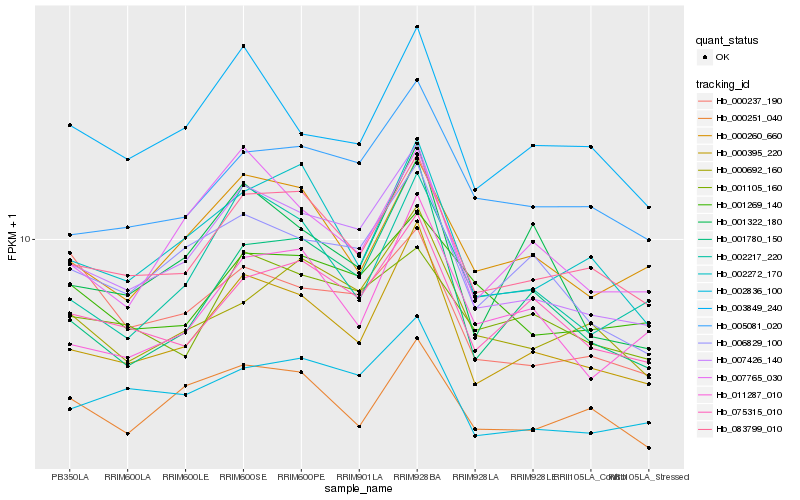
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007426_140 |
0.0 |
- |
- |
ornithine aminotransferase, putative [Ricinus communis] |
| 2 |
Hb_000395_220 |
0.0648711933 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 3 |
Hb_083799_010 |
0.0840751781 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 4 |
Hb_002836_100 |
0.0849490639 |
- |
- |
PREDICTED: nudix hydrolase 3 isoform X1 [Jatropha curcas] |
| 5 |
Hb_006829_100 |
0.0863955709 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_075315_010 |
0.1043856171 |
- |
- |
ornithine aminotransferase [Camellia sinensis] |
| 7 |
Hb_003849_240 |
0.1056829501 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 8 |
Hb_005081_020 |
0.1062972511 |
- |
- |
PREDICTED: ninja-family protein mc410 [Jatropha curcas] |
| 9 |
Hb_002272_170 |
0.1068812207 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase-like protein 1 [Jatropha curcas] |
| 10 |
Hb_007765_030 |
0.1072647932 |
- |
- |
PREDICTED: neutral ceramidase [Jatropha curcas] |
| 11 |
Hb_002217_220 |
0.1079670519 |
- |
- |
hypothetical protein POPTR_0010s15930g [Populus trichocarpa] |
| 12 |
Hb_001780_150 |
0.1115719919 |
- |
- |
ceramidase, putative [Ricinus communis] |
| 13 |
Hb_000692_160 |
0.1137284996 |
- |
- |
Transportin 1 isoform 1 [Theobroma cacao] |
| 14 |
Hb_000260_660 |
0.1147249173 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001105_160 |
0.1165420187 |
- |
- |
PREDICTED: protein phosphatase 2C 32 [Jatropha curcas] |
| 16 |
Hb_001322_180 |
0.1170362642 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 26 [Jatropha curcas] |
| 17 |
Hb_001269_140 |
0.1175101809 |
- |
- |
PREDICTED: nuclear pore complex protein NUP96 [Jatropha curcas] |
| 18 |
Hb_011287_010 |
0.1194459367 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 19 |
Hb_000251_040 |
0.1213058682 |
- |
- |
PREDICTED: DNA polymerase alpha catalytic subunit isoform X1 [Jatropha curcas] |
| 20 |
Hb_000237_190 |
0.1213125177 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SNF2 domain-containing protein CLASSY 3 isoform X1 [Jatropha curcas] |