| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
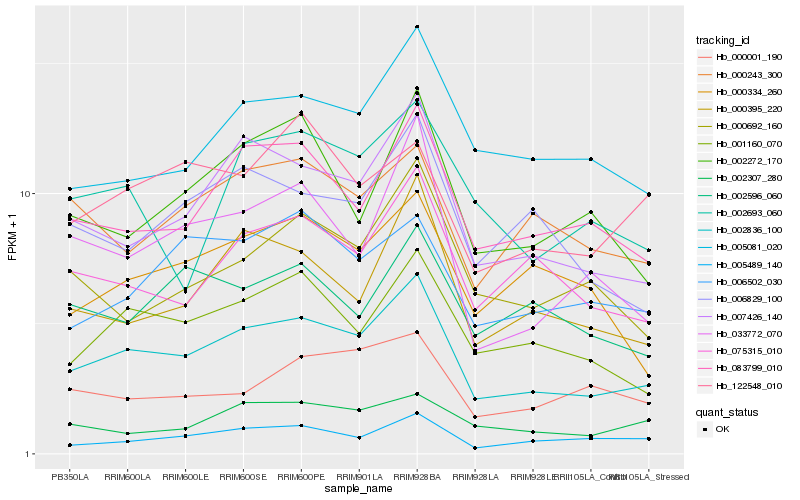
Hb_002836_100 |
0.0 |
- |
- |
PREDICTED: nudix hydrolase 3 isoform X1 [Jatropha curcas] |
| 2 |
Hb_007426_140 |
0.0849490639 |
- |
- |
ornithine aminotransferase, putative [Ricinus communis] |
| 3 |
Hb_083799_010 |
0.1023966856 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 4 |
Hb_001160_070 |
0.104560942 |
- |
- |
PREDICTED: uncharacterized protein LOC105632163 [Jatropha curcas] |
| 5 |
Hb_000395_220 |
0.1090327728 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 6 |
Hb_075315_010 |
0.1128398179 |
- |
- |
ornithine aminotransferase [Camellia sinensis] |
| 7 |
Hb_033772_070 |
0.1134875019 |
- |
- |
PREDICTED: uncharacterized protein LOC105633681 [Jatropha curcas] |
| 8 |
Hb_005489_140 |
0.1164974638 |
- |
- |
PREDICTED: maspardin [Jatropha curcas] |
| 9 |
Hb_005081_020 |
0.1193403616 |
- |
- |
PREDICTED: ninja-family protein mc410 [Jatropha curcas] |
| 10 |
Hb_006829_100 |
0.1198031069 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002272_170 |
0.1214268983 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase-like protein 1 [Jatropha curcas] |
| 12 |
Hb_006502_030 |
0.1233273338 |
- |
- |
PREDICTED: probable kinetochore protein ndc80 [Jatropha curcas] |
| 13 |
Hb_000692_160 |
0.1260896016 |
- |
- |
Transportin 1 isoform 1 [Theobroma cacao] |
| 14 |
Hb_000243_300 |
0.1278457182 |
- |
- |
catalytic, putative [Ricinus communis] |
| 15 |
Hb_000334_260 |
0.1293112206 |
- |
- |
carotenoid cleavage dioxygenase 1 [Manihot esculenta] |
| 16 |
Hb_002307_280 |
0.1302904142 |
- |
- |
Endonuclease/exonuclease/phosphatase family protein isoform 3 [Theobroma cacao] |
| 17 |
Hb_122548_010 |
0.1317304215 |
- |
- |
acyl-CoA oxidase, putative [Ricinus communis] |
| 18 |
Hb_000001_190 |
0.1324024404 |
- |
- |
hypothetical protein CISIN_1g0082522mg, partial [Citrus sinensis] |
| 19 |
Hb_002693_060 |
0.1329760656 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 20 |
Hb_002596_060 |
0.1346834825 |
- |
- |
conserved hypothetical protein [Ricinus communis] |