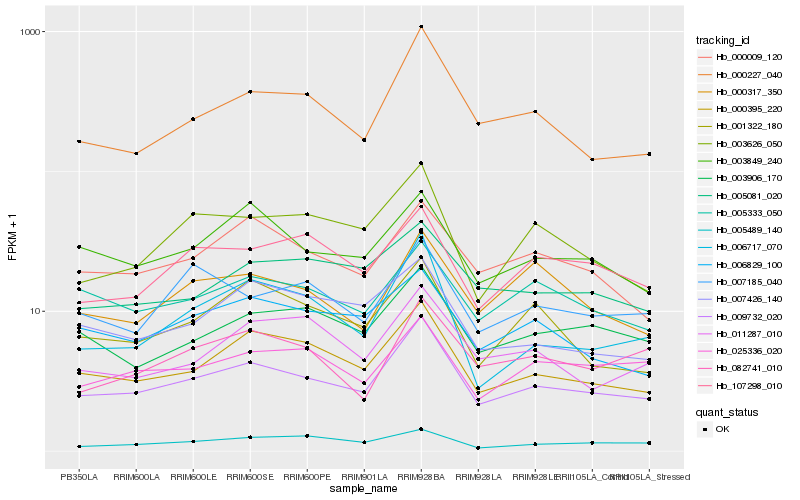
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009732_020 |
0.0 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Jatropha curcas] |
| 2 |
Hb_000395_220 |
0.0830918341 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 3 |
Hb_000227_040 |
0.0896579253 |
- |
- |
Stem-specific protein TSJT1, putative [Ricinus communis] |
| 4 |
Hb_003906_170 |
0.1075293938 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 5 |
Hb_006717_070 |
0.1108213664 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor CPC-like [Jatropha curcas] |
| 6 |
Hb_005333_050 |
0.1150886673 |
- |
- |
PREDICTED: glucose-6-phosphate isomerase 1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_003849_240 |
0.1152266393 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 8 |
Hb_000009_120 |
0.1164999156 |
- |
- |
PREDICTED: probable Xaa-Pro aminopeptidase P isoform X1 [Jatropha curcas] |
| 9 |
Hb_003626_050 |
0.1169447789 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 10 |
Hb_006829_100 |
0.1189609126 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_025336_020 |
0.1191105327 |
- |
- |
PREDICTED: PP2A regulatory subunit TAP46 [Jatropha curcas] |
| 12 |
Hb_000317_350 |
0.1226734261 |
- |
- |
PREDICTED: protein SUPPRESSOR OF GENE SILENCING 3 [Jatropha curcas] |
| 13 |
Hb_005081_020 |
0.1233246959 |
- |
- |
PREDICTED: ninja-family protein mc410 [Jatropha curcas] |
| 14 |
Hb_007185_040 |
0.1237924615 |
- |
- |
PREDICTED: pyrroline-5-carboxylate reductase [Jatropha curcas] |
| 15 |
Hb_001322_180 |
0.1238819865 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 26 [Jatropha curcas] |
| 16 |
Hb_107298_010 |
0.1239478037 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF2A isoform X2 [Jatropha curcas] |
| 17 |
Hb_082741_010 |
0.1243287772 |
- |
- |
PREDICTED: uncharacterized protein LOC105649930 [Jatropha curcas] |
| 18 |
Hb_007426_140 |
0.1248584325 |
- |
- |
ornithine aminotransferase, putative [Ricinus communis] |
| 19 |
Hb_011287_010 |
0.1252176319 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 20 |
Hb_005489_140 |
0.1254249673 |
- |
- |
PREDICTED: maspardin [Jatropha curcas] |