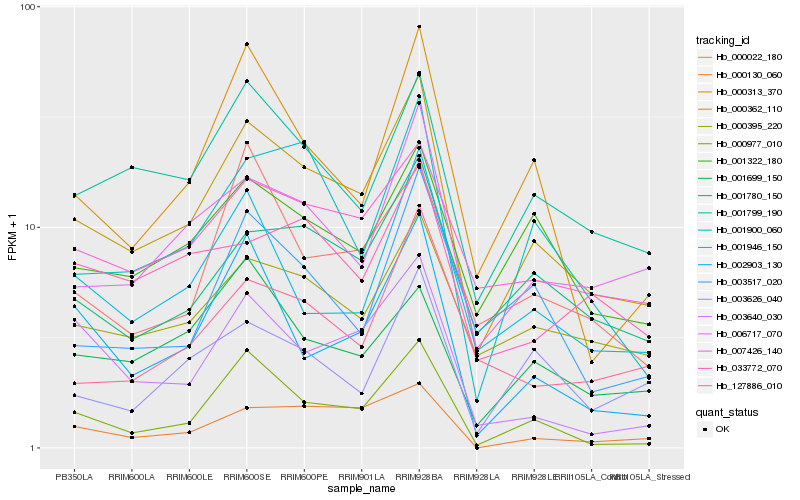
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000362_110 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104612097 isoform X2 [Nelumbo nucifera] |
| 2 |
Hb_006717_070 |
0.1234403828 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor CPC-like [Jatropha curcas] |
| 3 |
Hb_000395_220 |
0.1249188271 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 4 |
Hb_001780_150 |
0.1260032017 |
- |
- |
ceramidase, putative [Ricinus communis] |
| 5 |
Hb_001799_190 |
0.1265370671 |
- |
- |
PREDICTED: uncharacterized protein LOC105643221 [Jatropha curcas] |
| 6 |
Hb_007426_140 |
0.1282074337 |
- |
- |
ornithine aminotransferase, putative [Ricinus communis] |
| 7 |
Hb_000977_010 |
0.1352502939 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 8 |
Hb_001900_060 |
0.1360516959 |
- |
- |
PREDICTED: neutral alpha-glucosidase C [Jatropha curcas] |
| 9 |
Hb_001322_180 |
0.1369000419 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 26 [Jatropha curcas] |
| 10 |
Hb_001946_150 |
0.1377701089 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000313_370 |
0.1422216911 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 12 |
Hb_000130_060 |
0.1447185705 |
- |
- |
PREDICTED: uncharacterized protein LOC105641067 [Jatropha curcas] |
| 13 |
Hb_002903_130 |
0.1524044473 |
- |
- |
epoxide hydrolase, putative [Ricinus communis] |
| 14 |
Hb_000022_180 |
0.1549143872 |
- |
- |
PREDICTED: RING finger and CHY zinc finger domain-containing protein 1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_003517_020 |
0.1552742807 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter C family member 3-like [Jatropha curcas] |
| 16 |
Hb_127886_010 |
0.1556792248 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 17 |
Hb_003626_040 |
0.1559634194 |
- |
- |
PREDICTED: probable 2-oxoglutarate-dependent dioxygenase At3g49630 isoform X2 [Jatropha curcas] |
| 18 |
Hb_003640_030 |
0.157839304 |
- |
- |
PREDICTED: uncharacterized protein LOC105647345 isoform X1 [Jatropha curcas] |
| 19 |
Hb_033772_070 |
0.1588795642 |
- |
- |
PREDICTED: uncharacterized protein LOC105633681 [Jatropha curcas] |
| 20 |
Hb_001699_150 |
0.1597290824 |
- |
- |
hypothetical protein POPTR_0009s13450g [Populus trichocarpa] |