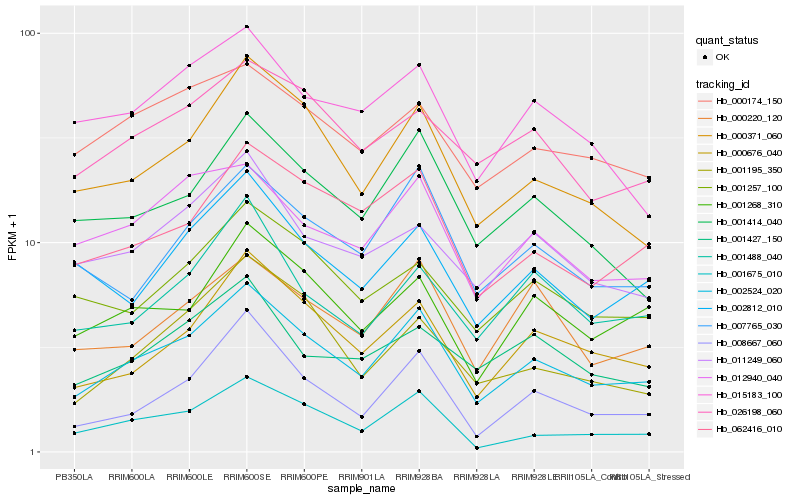
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002524_020 |
0.0 |
- |
- |
glycerophosphoryl diester phosphodiesterase, putative [Ricinus communis] |
| 2 |
Hb_000371_060 |
0.0867383668 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 1 protein, putative [Ricinus communis] |
| 3 |
Hb_000676_040 |
0.0879015597 |
- |
- |
hypothetical protein POPTR_0007s13320g [Populus trichocarpa] |
| 4 |
Hb_062416_010 |
0.0980604045 |
- |
- |
PREDICTED: UPF0202 protein At1g10490 [Jatropha curcas] |
| 5 |
Hb_001414_040 |
0.1001000002 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 1-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_001268_310 |
0.100783158 |
- |
- |
- |
| 7 |
Hb_007765_030 |
0.1019964138 |
- |
- |
PREDICTED: neutral ceramidase [Jatropha curcas] |
| 8 |
Hb_000220_120 |
0.1032985775 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 isoform X2 [Jatropha curcas] |
| 9 |
Hb_012940_040 |
0.106190652 |
- |
- |
Splicing factor, arginine/serine-rich, putative [Ricinus communis] |
| 10 |
Hb_001675_010 |
0.1068247026 |
- |
- |
hypothetical protein TRIUR3_09961 [Triticum urartu] |
| 11 |
Hb_008667_060 |
0.1079297915 |
- |
- |
Magnesium and cobalt efflux protein corC, putative [Ricinus communis] |
| 12 |
Hb_001257_100 |
0.1085068902 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor E2FB isoform X1 [Jatropha curcas] |
| 13 |
Hb_001488_040 |
0.1090114479 |
- |
- |
PREDICTED: nipped-B-like protein A [Jatropha curcas] |
| 14 |
Hb_026198_060 |
0.1092460847 |
- |
- |
PREDICTED: calcium-dependent protein kinase 11 [Jatropha curcas] |
| 15 |
Hb_002812_010 |
0.1103168809 |
- |
- |
PREDICTED: chitin elicitor receptor kinase 1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_011249_060 |
0.1124207364 |
- |
- |
hypothetical protein POPTR_0018s12600g [Populus trichocarpa] |
| 17 |
Hb_001195_350 |
0.1125121325 |
- |
- |
hypothetical protein JCGZ_04423 [Jatropha curcas] |
| 18 |
Hb_001427_150 |
0.1132493043 |
- |
- |
PREDICTED: uncharacterized protein LOC105637790 isoform X1 [Jatropha curcas] |
| 19 |
Hb_015183_100 |
0.1146972775 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360 [Jatropha curcas] |
| 20 |
Hb_000174_150 |
0.1150984674 |
- |
- |
stearoyl-acyl-carrier protein desaturase [Ricinus communis] |