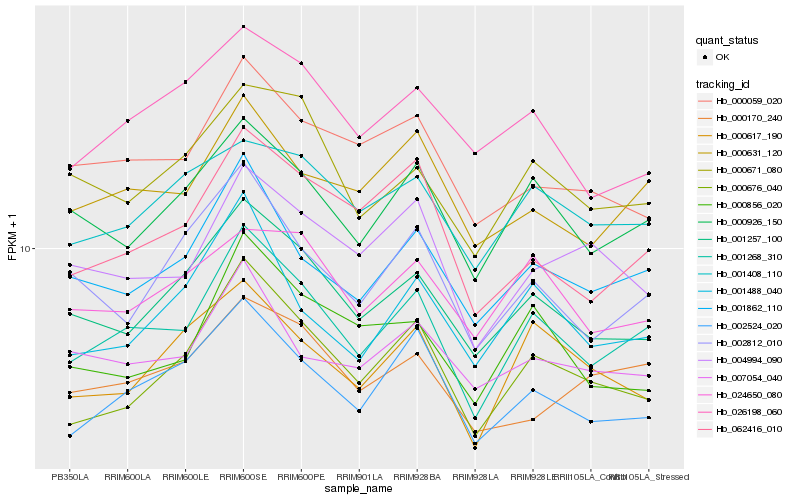
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001268_310 |
0.0 |
- |
- |
- |
| 2 |
Hb_001862_110 |
0.0909287738 |
- |
- |
PREDICTED: U-box domain-containing protein 4 [Jatropha curcas] |
| 3 |
Hb_000926_150 |
0.0926937819 |
- |
- |
PREDICTED: splicing factor, suppressor of white-apricot homolog isoform X1 [Jatropha curcas] |
| 4 |
Hb_001257_100 |
0.0966228971 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor E2FB isoform X1 [Jatropha curcas] |
| 5 |
Hb_002524_020 |
0.100783158 |
- |
- |
glycerophosphoryl diester phosphodiesterase, putative [Ricinus communis] |
| 6 |
Hb_000631_120 |
0.1010478304 |
- |
- |
hypothetical protein CISIN_1g0431902mg, partial [Citrus sinensis] |
| 7 |
Hb_000676_040 |
0.1019577143 |
- |
- |
hypothetical protein POPTR_0007s13320g [Populus trichocarpa] |
| 8 |
Hb_062416_010 |
0.106939822 |
- |
- |
PREDICTED: UPF0202 protein At1g10490 [Jatropha curcas] |
| 9 |
Hb_000671_080 |
0.1088346064 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase-like 4 [Jatropha curcas] |
| 10 |
Hb_001488_040 |
0.1114217143 |
- |
- |
PREDICTED: nipped-B-like protein A [Jatropha curcas] |
| 11 |
Hb_002812_010 |
0.1137550766 |
- |
- |
PREDICTED: chitin elicitor receptor kinase 1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000059_020 |
0.1169141994 |
- |
- |
PREDICTED: F-box protein SKP2A [Jatropha curcas] |
| 13 |
Hb_004994_090 |
0.1171778829 |
- |
- |
PREDICTED: uncharacterized protein LOC105643033 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000856_020 |
0.1177121126 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 19 [Jatropha curcas] |
| 15 |
Hb_001408_110 |
0.1178113498 |
- |
- |
hypothetical protein EUGRSUZ_I02762 [Eucalyptus grandis] |
| 16 |
Hb_007054_040 |
0.1179031452 |
- |
- |
PREDICTED: protein RST1 [Jatropha curcas] |
| 17 |
Hb_000170_240 |
0.1187647461 |
- |
- |
PREDICTED: auxilin-related protein 1 [Jatropha curcas] |
| 18 |
Hb_000617_190 |
0.1187835509 |
- |
- |
PREDICTED: heparan-alpha-glucosaminide N-acetyltransferase [Jatropha curcas] |
| 19 |
Hb_026198_060 |
0.1188803282 |
- |
- |
PREDICTED: calcium-dependent protein kinase 11 [Jatropha curcas] |
| 20 |
Hb_024650_080 |
0.1189526059 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g09030 [Jatropha curcas] |