| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
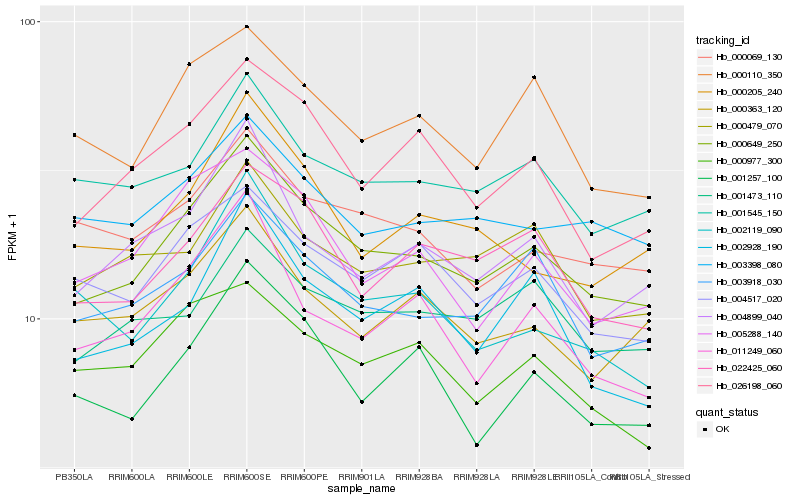
Hb_000649_250 |
0.0 |
transcription factor |
TF Family: C2H2 |
PREDICTED: UBX domain-containing protein 6 [Jatropha curcas] |
| 2 |
Hb_003918_030 |
0.0703547175 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 1C-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_026198_060 |
0.0708999664 |
- |
- |
PREDICTED: calcium-dependent protein kinase 11 [Jatropha curcas] |
| 4 |
Hb_004899_040 |
0.0730541721 |
transcription factor |
TF Family: ARF |
Auxin response factor [Medicago truncatula] |
| 5 |
Hb_022425_060 |
0.0747899 |
- |
- |
PREDICTED: protein phosphatase 2C and cyclic nucleotide-binding/kinase domain-containing protein isoform X1 [Jatropha curcas] |
| 6 |
Hb_004517_020 |
0.0752678835 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2B isoform X1 [Jatropha curcas] |
| 7 |
Hb_001257_100 |
0.0754203923 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor E2FB isoform X1 [Jatropha curcas] |
| 8 |
Hb_000110_350 |
0.0763582713 |
- |
- |
PREDICTED: ubiquitin receptor RAD23c-like isoform X2 [Gossypium raimondii] |
| 9 |
Hb_005288_140 |
0.0763927116 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A regulatory subunit B''beta-like [Jatropha curcas] |
| 10 |
Hb_001545_150 |
0.0780877879 |
transcription factor |
TF Family: NAC |
PREDICTED: uncharacterized protein LOC105635527 [Jatropha curcas] |
| 11 |
Hb_000069_130 |
0.0797748573 |
- |
- |
PREDICTED: uncharacterized protein LOC105642829 isoform X1 [Jatropha curcas] |
| 12 |
Hb_003398_080 |
0.0802425054 |
- |
- |
PREDICTED: telomere repeat-binding protein 4 isoform X3 [Jatropha curcas] |
| 13 |
Hb_011249_060 |
0.0808693705 |
- |
- |
hypothetical protein POPTR_0018s12600g [Populus trichocarpa] |
| 14 |
Hb_000205_240 |
0.0816281335 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 15 |
Hb_002928_190 |
0.0825108959 |
transcription factor |
TF Family: RWP-RK |
PREDICTED: protein NLP4 [Jatropha curcas] |
| 16 |
Hb_000363_120 |
0.083550762 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 20 [Jatropha curcas] |
| 17 |
Hb_001473_110 |
0.0844877421 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 13C-like [Jatropha curcas] |
| 18 |
Hb_002119_090 |
0.0846589608 |
- |
- |
PREDICTED: protein OBERON 3 [Jatropha curcas] |
| 19 |
Hb_000977_300 |
0.0848019025 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable E3 ubiquitin-protein ligase ARI8 [Malus domestica] |
| 20 |
Hb_000479_070 |
0.0852990065 |
- |
- |
PREDICTED: uncharacterized protein LOC105644141 [Jatropha curcas] |